International Journal of Nanomaterials, Nanotechnology and Nanomedicine
Study of silver nanoparticle effects on some molecular responses and metabolic pathways of Phytophthora parasitica
Shaheen Bibi1, Jose C Huguet-Tapia1, Zunaira Afzal Naveed1,2, Ashraf SA El-Sayed2#, Sujan Timilsina1, Jeffrey B Jones1* and Gul Shad Ali2
2Mid-Florida Research and Education Center, Department of Plant Pathology, University of Florida/Institute of Food and Agricultural Sciences, 2725 Binion Rd, Apopka, FL 32703, USA
#Botany and Microbiology Department, Faculty of Science, Zagazig University, Egypt
Cite this as
Bibi S, Huguet-Tapia JC, Naveed ZA, El-Sayed ASA, Jones JB, et al. (2021) Study of silver nanoparticle effects on some molecular responses and metabolic pathways of Phytophthora parasitica. Int J Nanomater Nanotechnol Nanomed 7(2): 047-056. DOI: 10.17352/2455-3492.000046Phytophthora parasitica is a devastating plant pathogen that has a wide host range. As a new approach, silver nanoparticles (AgNPs) were assessed to control it. Previously AgNPs were shown to inhibit mycelial growth, zoospore production and germination, and germ tube elongation. However, the mechanism(s) of bioactivity of AgNPs on changes in metabolic patterns in P. parasiticahas not been completely resolved. P. parasiticawas exposed to AgNPs at several time points, RNA-Sequences were extracted, and then selected key genes in several of the pathways were verified for the level of expression using qRT-PCR. For RNA-Seq, the reads were mapped to the P. parasiticagenome INRA 310.3. Principle component analysis illustrated low variation among biological replicates and emphasized the reproducibility of results. In this paper, seven candidate genes identified by RNA-Seq were evaluated for their expression using qRT-PCR. Interestingly, qRT-PCR confirmed that the genes that were significantly altered were found to be involved in major cellular pathways such as glutathione metabolism and ribosome biogenesis. qRT-PCR also revealed heat shock protein, ABC transporter, and Glutathione-S-Transferase (GST) genes were significantly affected. These genes are related to oxidative stress. Expression of GST and heat-shock protein significantly increased at 1 hr post-exposure to AgNPs, indicating a rapid response. The analysis based on qRT-PCR makes it evident that key genes involved in many major pathways were significantly altered on exposure to AgNPs.
Abbreviations
DEG: Differentially Expressed Gene; HC: Hierarchical Clustering; GO: Gene Ontology; PCA: Principle Component Analysis; RIN: Integrity Numbers; WebMGA: Web Services for Metagenomic Analysis; qRT-PCR: Real Time Quantitative Reverse Transcription PCR
Introduction
The genus Phytophthora includes more than 120 species, causing many devastating diseases to plants, natural ecosystems, and crops worldwide [1]. Phytophthora infestans is the most notable species, and was the primary cause of the destruction of potato monoculture in Europe, contributing to a great famine that resulted in the reduction of the total Irish population by approximately 20% in the 1840s [2]. More research has been focused mainly on P. infestans and P. sojae. P. infestans infects potato and tomato and is a foliar pathogen [3], while P. sojae causes stem and root rot of soybean [4]. P. parasiticais a typical root pathogen that infects over 255 plant genera in 90 families [5]. The hosts range from field crops [6] to forest trees [7] in natural habitats [8]. P. parasiticais a worldwide problem causing black shank disease of tobacco [9] and root rot and gummosis of citrus species [10].
Oomycetes which include the genus Phytophthora, are coenocytic [11] and have notable differences in cell wall composition compared to fungi. Oomycetes mainly consist of β-1,3-glucan and β-1,6-glucan polymers and cellulose, whereas fungal cell walls are mainly composed of chitin [12]. This difference makes oomycetes challenging to control as most of the fungicides target sterol and chitin synthesis which is absent in oomycetes (8). The phenylamide fungicide, metalaxyl, used against oomycete pathogens, targets and inhibits RNA polymerase 1 [13]. However, there has been development of resistance reported against metalaxyl in closely related P. infestans populations [14]. Phytophthora species are well known to develop resistance to chemicals rapidly [15-19].
In order to manage pathogens, new strategies are currently being explored and evaluated with the aid of genomics and transcriptomics. Nanoparticles have gained a lot of interest in recent years for use in disease control based on their biological, chemical, and physical properties. They have been used in disease management based on these unique characteristics [20]. Silver nanoparticles (AgNPs) have been used to control fungal pathogens because of their multiple modes of inhibition to microorganisms [21]. There are several studies reporting the efficacy of AgNPs for controlling diverse pathogens, including bacteria [22] and fungi [23]. Antibacterial and antifungal activities of AgNPs have been shown to have great potential in controlling bacteria and spore-producing fungi, with less toxicity to the ecosystem [24]. AgNPs affect a broad range of metabolic processes and have been shown to have multiple modes of action that make pathogens less prone to developing resistance [25]. AgNPs act in three significant ways to inhibit microorganisms: they interact with the cell membranes or cell walls, bind to DNA [26], and bind to proteins [27]. Silver ions have been reported to interact with thiols, amides, carboxylates, and hydroxyl groups [28-30]. Such interactions can result in targeting membrane-bound cytoplasmic enzymes and proteins as well as DNA. These chemical interactions have been reported to cause changes in the structure of bacterial membranes [26] inhibit respiration [31], and increase reactive oxygen species [30].
Silver nanoparticles (AgNPs) have shown promise for managing P. parasiticabecause of properties attributed to their size and large ratio of surface area to volume [32]. In a study by Ali, et al. [32], they demonstrated that these Artemisia absinthium based AgNPs inhibited the growth of P. parasiticaat various developmental stages. AgNPs exhibited a potent inhibitory effect on the growth of mycelia and zoospore germination. The mycelial growth was reported to be inhibited strongly after 12 hours of incubation with AgNPs. Although several studies have outlined mode of action of AgNPs in bacterial [33,34] and fungal pathogens [23], the precise mode of action of AgNPs against P. parasiticais not fully understood. This study aimed to elucidate the changes in the cellular metabolism induced by AgNP toxicity by quantifying and studying genes associated with the pathways of P. parasitica.
Results and discussion
Exploratory transcriptome analysis of P. parasitica transcriptome in response to AgNPs
The exploratory expression studies we used provided a general overview of the impact of AgNPs on P. parasiticaand provided targets for qRT-PCR. Clean reads from all fourteen libraries were mapped to the P. parasiticagenome INRA 310.3 (AGFV00000000.2) ("(dataset) Broad Institute [35] Phytophthora parasitica INRA-310 Genome sequencing and assembly. National Center for Biotechnology Information,"). All the samples showed overall mapping coverage of 74-85% to the P. parasiticagenome. Details of the mapping and sequences are given in Table S1. Dramatic differences in P. parasiticatranscriptome were observed in the samples treated with AgNPs compared to the control samples.
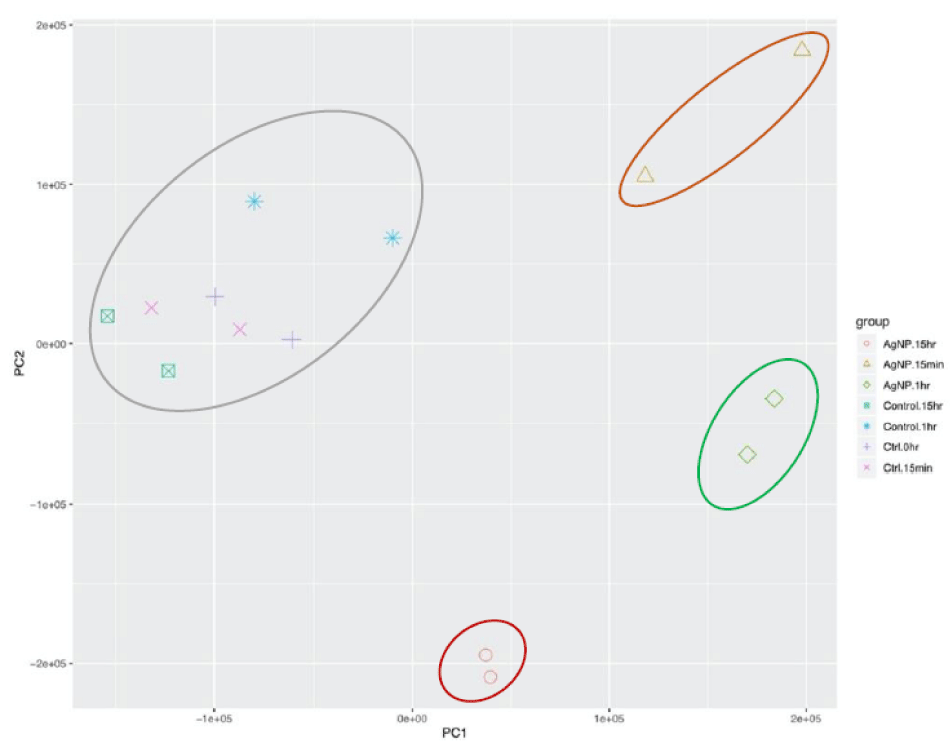
To check the variation among all the samples and the quality of our data, we conducted principal component analysis (PCA) (Figure 1A) and hierarchical clustering (HC) (Figure S1). The normalized differences in expression patterns were used to compute a distance matrix. Distances among samples and replicates can be displayed in two dimensions. PCA discriminated between treatments and replicates based on 76% of the variance (PC1 accounted for 53% of the variance and PC2 for 23% of the variance). Clustering results indicated low variation among the treatment replicates, indicating reproducibility of the biological replicates. P. parasiticatreated with AgNPs showed considerable separation compared to the mock-treated control samples at all time points, indicating transcriptome changes in response to the application of AgNPs (Figure 1A). Furthermore, the biological reps for each time point following AgNPs exposure clustered together but were separated from those at the other time points.
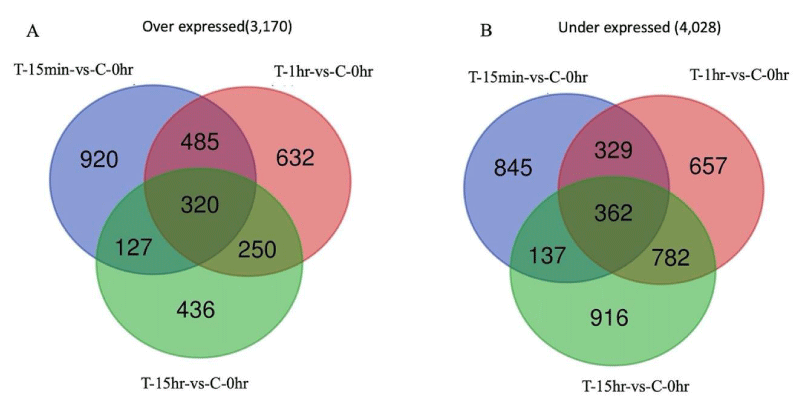
Genes that were altered significantly in their expression in treated samples were identified for each time point at 15 min, 1 hr, and 15 hr (Table S2). The highest number of differentially expressed genes (DEGs), 3818, was found in AgNP-treated samples at 1 hr vs mock-treated control samples at 0 hr. We also compared the DEGs among AgNP-treated samples at different time points and found the highest number of DEGs among AgNP-treated samples at 15 hr vs. AgNP-treated samples at 15 min. Venn diagram analysis indicated a total of 4,028 genes were under expressed collectively in all treated samples at time points 15 min, 1 hr, and 15 hr compared to mock-treated control at 0 hr, while 3,170 genes were over expressed at these time points (Figure 1B). Gene expression comparisons with adjacent time points showed a total of 4,531 genes under expressed and 4,108 genes over expressed in samples treated with AgNPs (Figure S2). The genes at each time point in these Venn diagram analyses are provided in Table S3. We identified several processes that were downregulated, such as ribosome biogenesis, DNA replication, and respiration. The results of RNA-Seq give a general picture, and the results are broadly in line with previous studies [34,36] that have reported the same process to be significantly affected in response to AgNPs. Gene ontology (GO) enrichment analysis is frequently used to determine the biological activity of genes. The differentially expressed genes were mapped in terms of their GO analysis to obtain the possible effect of AgNPs. It was seen that GO categories such as metabolic processes and cellular processes were predominantly affected. Unlike fungicides and antibiotics, AgNPs appear to target several processes. Based on the preliminary RNA-Seq data, we identified genes associated with oxidative stress, which were altered in gene expression following exposure to AgNPs.
qRT-PCR indicates alteration of oxidative stress-related genes in exposure to AgNPs
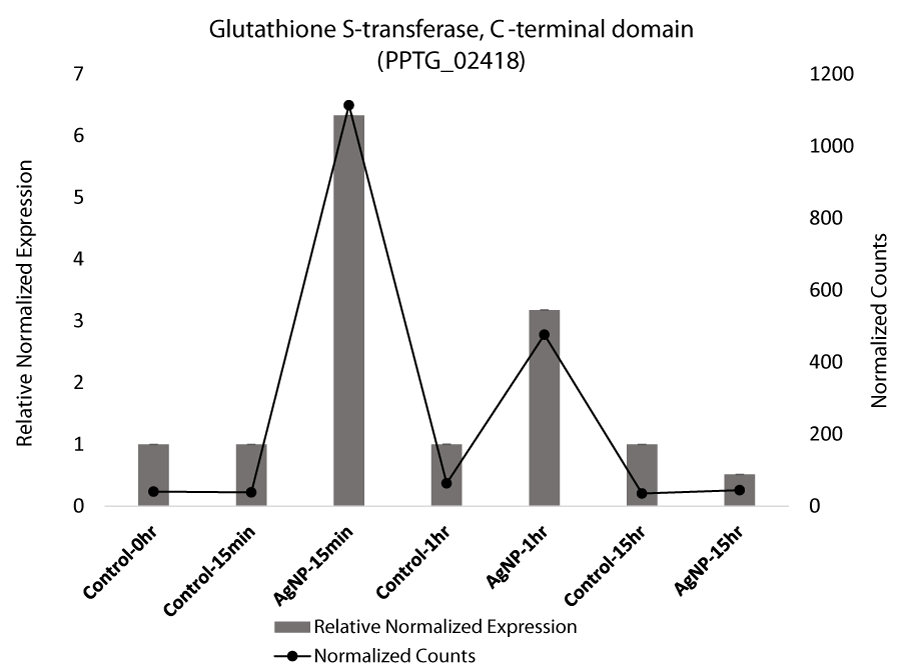
Glutathione S-transferase (GST), which is associated with stress in plants [37] and fungi [38] and which was identified in the exploratory transcriptome analyses, was targeted in qRT-PCR given that the transcript levels of GST were differentially expressed in samples that were treated with AgNPs compared to the mock-treated control. Based on qRT-PCR, GST was significantly upregulated at 15 min and 1 hr in AgNP-treated samples, but then the expression level decreased significantly at 15 hr (Figure 2 A). GST has been implicated in the detoxification of xenobiotic substances, and it was also indicated to be differentially regulated in our pathway analysis. The significant increase in GST gene expression in response to AgNPs, reveals the activation of this protein as part of the cellular antioxidant defense system. (Figure 2 A). Expression of glutathione peroxidase (EC 1.11.1.9), which is known to remove hydrogen peroxide, was also upregulated [33,39].
Glutathione activity is an important process involved in removing reactive oxygen species (ROS) generated endogenously during xenobiotic metabolism [40]. Glutathione redox is the marker for oxidative stress that plays a crucial role in resisting oxidative stress and maintaining cellular oxidation-reduction homeostasis [41]. Upregulation of GST in response to AgNPs and depletion of GSH suggests the imbalance caused in the cellular system and resulting ROS. The ROS can ultimately oxidize DNA and protein. Interestingly, AgNPs have been reported to act by generating ROS and depleting glutathione [39]. Oxidative stress refers to a state where GSH is depleted while oxidized glutathione accumulates [42]. In A. brassicicola, GST transcription was found to be elevated by heavy metals [43]. Bacterial GST genes are suggested to play an essential role in the degradation of xenobiotics and protect from oxidative stress. In a recent study on multidrug resistant P. aeruginosa, AgNPs had a strong bactericidal effect. Transmission electron microscopy showed AgNPs entering P. aeruginosa cells and impairing their structure and morphology by creating a disequilibrium of oxidation and reduction reactions and failing to eliminate excess reactive oxygen species [33,44]. Our analysis substantiates the previous reports where depletion of glutathione signifies oxidative stress [42]. An increased level of reduced glutathione is considered as a natural defense system in cells to overcome the metal generated oxidative stress [45,46]. The initial upregulation of the genes might be involved in defending the cells from the stress induced by AgNPs. With recent advances, the importance of GST and its role in the diverse cellular process was explored, confirming it to be a critical target for many inhibitors and drugs [47].
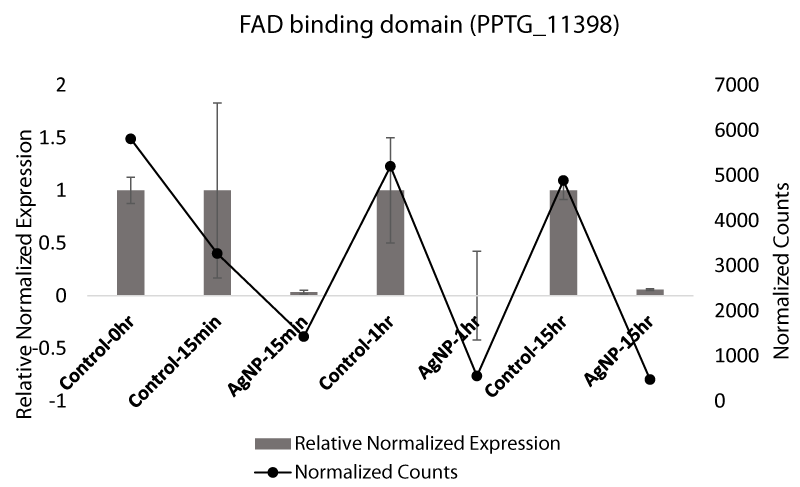
Flavin adenine dinucleotide (FAD)-binding domain, which was also identified in exploratory transcriptome analyses as differentially expressed, has previously been associated with stress in fungi [48]. We used qRT-PCR to measure the expression level of the FAD-binding domain in P. parasiticafollowing exposure to AgNPs. FAD-binding protein was expressed at 15 min post exposure and then was under expressed in all treated samples with AgNP-treated at 1 hr and 15 hr (Figure 2B). FAD-binding proteins are well known for their vital function in chromatin remodeling [49], DNA repair, protein folding [50], and dehydrogenation [51]. Exposure to AgNPs has enhanced their expression, likely to counteract the damage caused by the AgNPs. The qRT-PCR expression data support the gene expression data indicated by RNA-Seq analysis. FAD is involved in the dehydrogenation of many metabolites and has a role in oxidation [52].
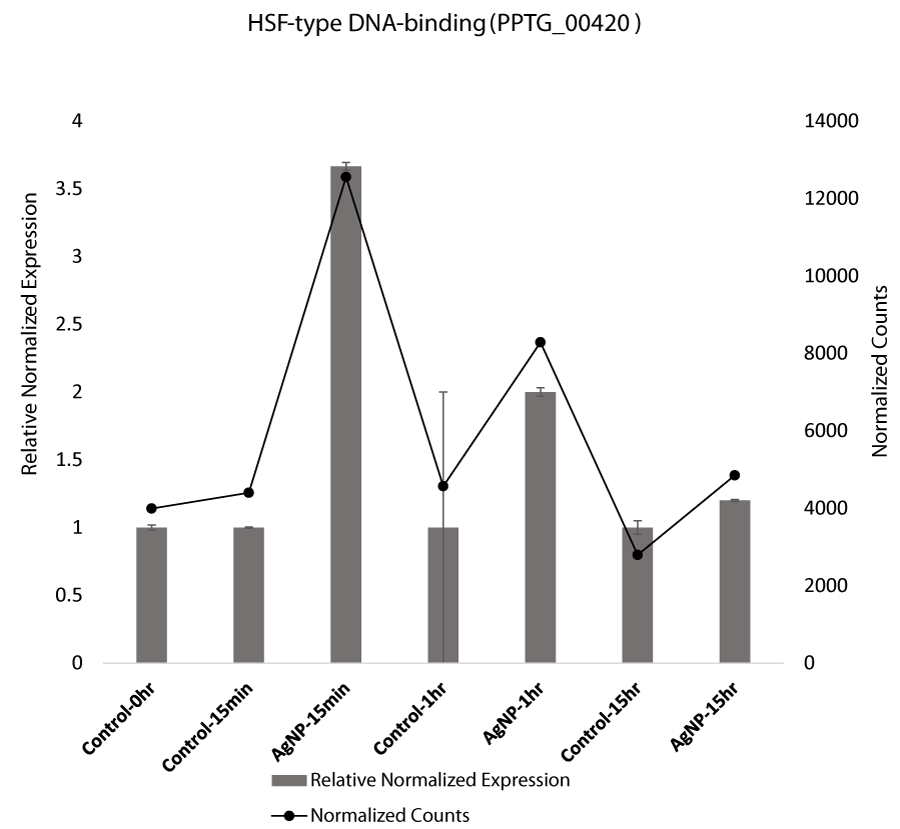
Stress at the cellular level is also induced in response to nanoparticles. Stress in cells is manifested by the production of stress related proteins, including heat shock proteins (Hsp), which are produced in cells that are exposed to extreme temperature, pH, nanoparticles, or heavy metals. Research indicated that heat shock protein expression increased fourfold in response to AgNPs [53]. Heat shock factors (HSFs) are activated in response to acute stress that binds the Hsp genes and mediates transcription [54]. Our qRT-PCR analysis confirmed that HSF-type DNA binding proteins were also over expressed at the initial exposure to AgNPs. (Figure 2C).
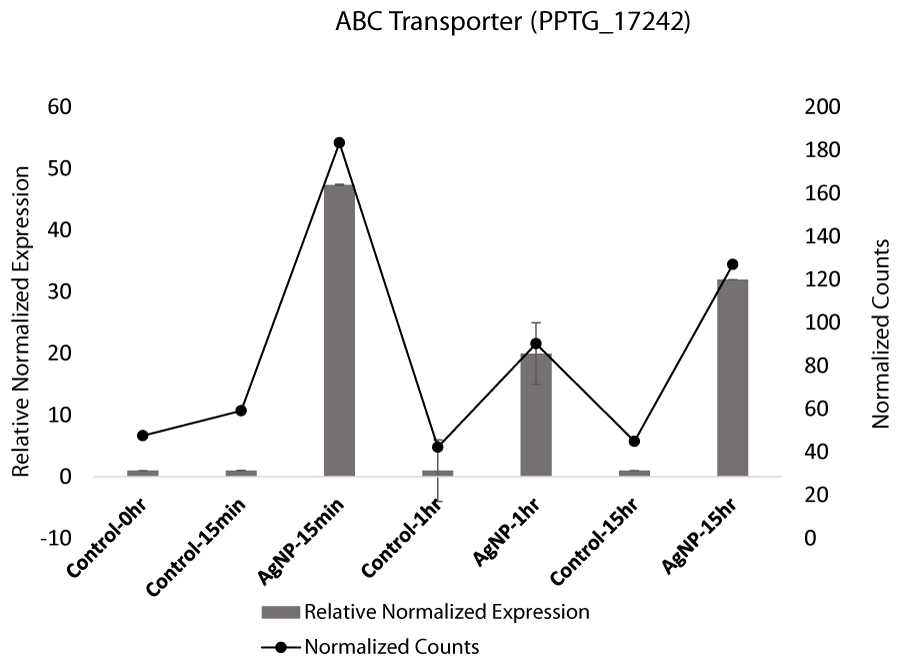
Initial high expression of ATP-binding cassette (ABC) transporter is also indicative of self-defense. ABC transporters exude a variety of substrates, including extracellular toxins from the cell [55,56]. However, research has indicated that AgNPs induce cell death and inhibit the efflux mechanism of the emergence of multidrug resistant (MDR) cancer cells [57]. ABC transporters have also been reported to be differentially regulated in response to AgNPs in P. aeruginosa [36]. In exploratory transcriptome analysis, expression of ABC transporter was increased at 15 min and decreased slightly later (Figure S10A). In contrast, the expression in control samples was not affected. The qRT-PCR analysis of ABC transporter gene (PPTG_17242) confirms the transcriptome analysis results where expression of the gene ABC transporter was upregulated at 15 min, 1 hr, and 15 hr compared to control samples. (Figure 2D). ABC transporters have also been reported to be involved in toxicant efflux and virulence [58-60]. These transporters serve as an apparatus for cell defense in many organisms, extruding various substrates [55]. The high expression in treated samples in response to AgNPs would indicate a stress response.
Alkyl hydroperoxide reductase subunit (AhpC) a subunit of thioredoxin, which has been reported to be an essential antioxidant and that has been associated with oxidative stress response as it directly reduces hyperoxides [61]. In this study it was found in exploratory transcriptome analysis to be downregulated two fold at 1 hr (Table S4). Downregulation of AhpC suggests a comparable decrease in antioxidant transcripts and increased oxidative stress.
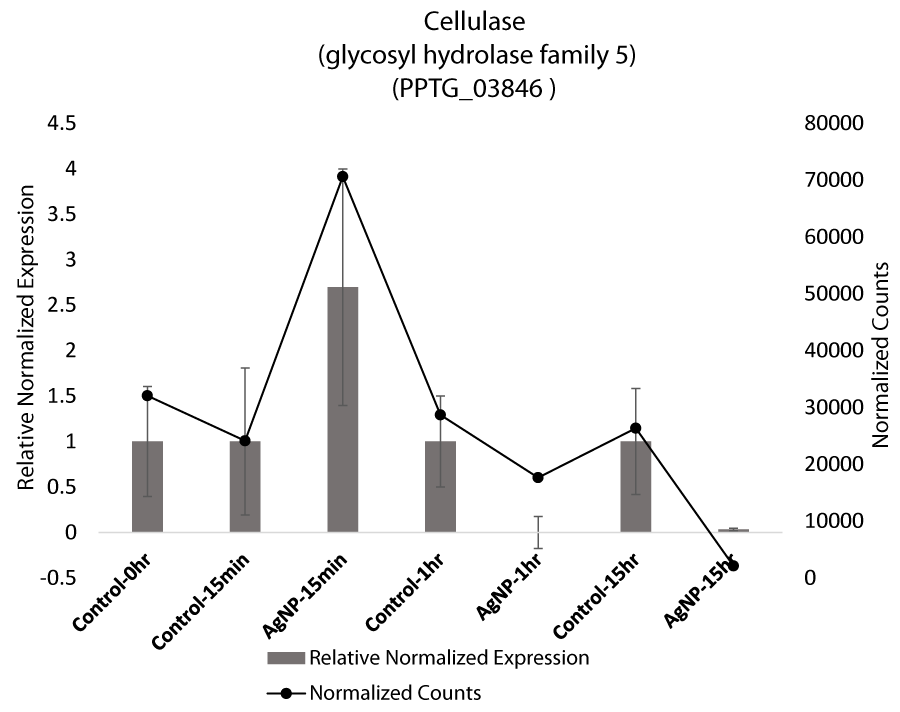
Apart from specific stress related genes, cellulase was also found to be differentially regulated based on qRT-PCR. Cellulases catalyze the degradation of cellulose present in cell walls including plants and Phytophthora. An increase in cellulase expression at 15 min and downregulation at 1 hr and 15 hr suggests that the stimulus of stress is perceived by the mycelia. (Figure 2E). Cellulase activity has also been reported to decrease with increasing abiotic stress of chromium. However, our qRT-PCR results for cellulases are not significant based on ANOVA (Table S5).
qRT-PCR reveals alteration of protein kinases in response to AgNPs
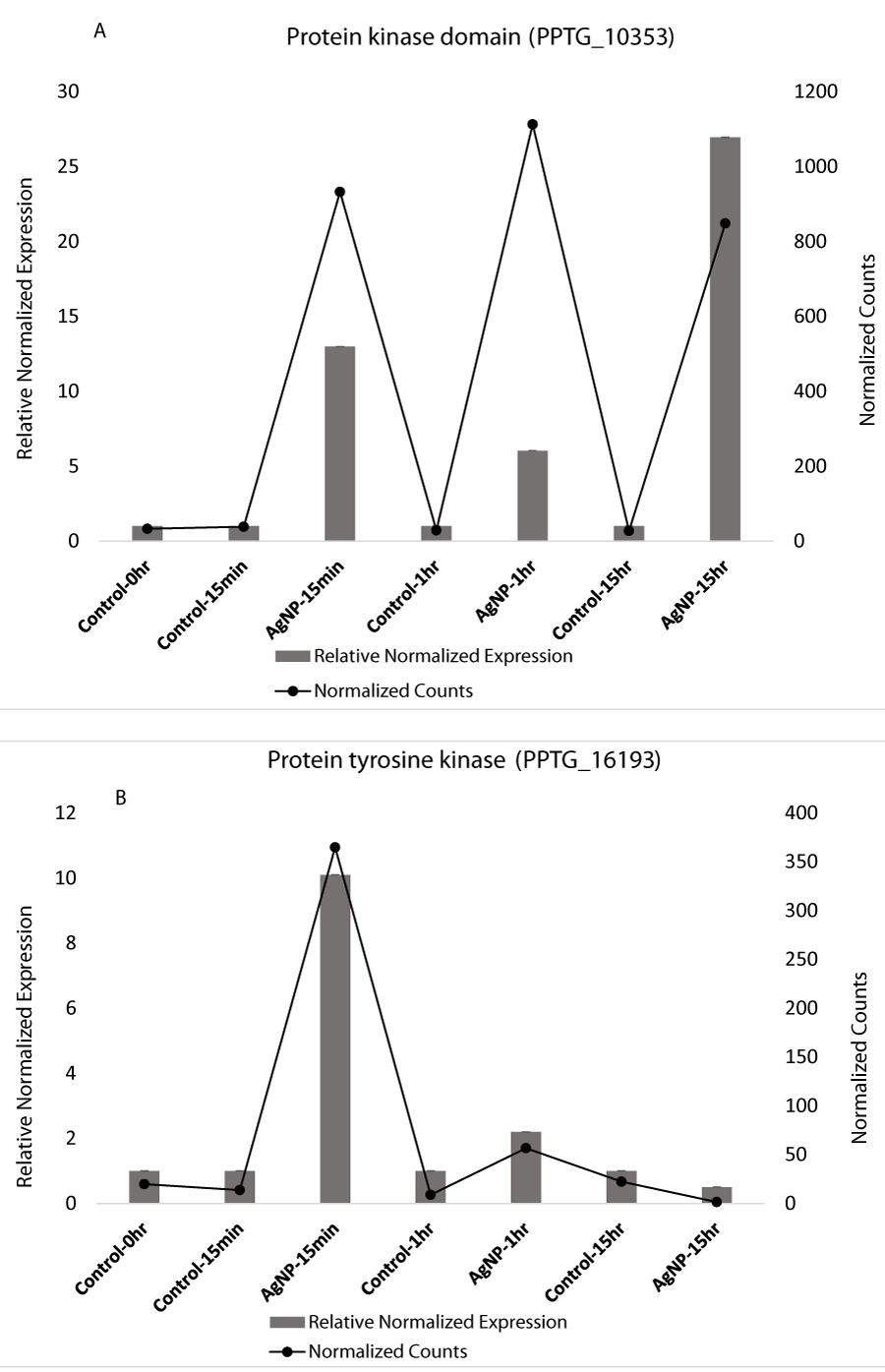
Apart from stress related genes, genes that contain protein kinase domains were also highly altered based on exploratory transcriptomic analysis and qRT-PCR data. Protein kinases play an important role in many cellular processes. The exploratory RNA-Seq analysis revealed that the expression of protein kinase domains significantly increased in treated samples on exposure to AgNPs (Table S2). We selected two kinases, a serine-threonine protein kinase catalytic domain (IPR000719) and a protein tyrosine kinase (IPR001245) from our transcriptomic analysis and observed their expression using qRT-PCR. We observed that protein kinase domain expression was high in all the treated samples while no change was observed in control samples (Figure 3A). qRT-PCR expression of protein tyrosine kinase indicated that expression of this gene increased at 15 min in treated samples while there was no observable difference in control samples. Dayem, et al. [62] also demonstrated that AgNPs enhance activation of protein kinases. Recently AgNPs have been reported to cause stress-induced apoptosis in the endoplasmic reticulum (ER). The damage is caused by differential regulation of various genes involved in ER stress and phosphorylation of protein kinases [63].
Protein kinase domain is structurally conserved and contains a catalytic function of typical protein kinases. Protein kinases catalyze the transfer of phosphate onto proteins and act as an on/off switch for many vital cellular processes including cell division, cell cycle, and metabolism [64]. Protein kinases play an essential role in intracellular signal communication and transcription as protein kinases regulate protein phosphorylation. Protein kinases regulate the protein phosphorylation by catalyzing the phosphorylation reaction where the gamma phosphate of ATP is transferred [65]. The ability of cells to perceive, recognize, and respond to physical and chemical stimuli is the key to their survival. Signal transduction is therefore important to react quickly and efficiently to these stimuli. A study on epidermal growth factor signal transduction indicated reduction of phosphorylation on exposure to silver nanoparticles and caused interference in signaling [66]. Perception of signals leads to certain modifications in the cell such as posttranslational modifications including phosphorylation and plays crucial roles in regulation of key cellular activities including protein binding affinity, stability, and activity [67]. Protein phosphorylation is regulated by protein kinases and kinases play an important role in signaling, cell division, and gene transcription [68]. Protein kinases have also been involved in non-catalytic process such as survival [69]. As the role of protein kinases is involved in several cellular processes, the differential expression on exposure to AgNPs results in changes in many protein activities which could be detrimental to the entire cellular machinery.
Pathways affected in response to AgNPs
Differential expression of 28 ribosomal biogenesis genes in response to AgNPs was an interesting observation, which suggests that ribosomes in P. parasitica could be a major target for AgNPs. Ribosome biogenesis involves maturation of ribosomal rRNAs and assembly into ribosomal subunits [70].
Ribosomes function at the heart of translation machinery for converting mRNA into protein and plays a vital role in cell growth in eukaryotes [71]. Ribosome biogenesis is highly conserved among eukaryotes and initial steps include rRNA precursors and transcription and assembly of factors and ribosomal proteins. In the eukaryotic cell, ribosome biogenesis requires more than 250 non- ribosomal assembly factors, of which many are vital. Despite many potential targets, only a few chemical inhibitors have been reported so far [72]. From pathway analysis, we observed that the genes that have a role in methylation and pseudouridylation were downregulated in AgNPs treated samples. These play important roles in modification of RNA (Figure S3). Our results indicated that AgNPs work as an inhibitory agent by interfering with ribosome biogenesis pathways by altering the expression of the genes essential for assembly of 90s pre-ribosome and affecting maturation of 40S and 60S (Figure S3).
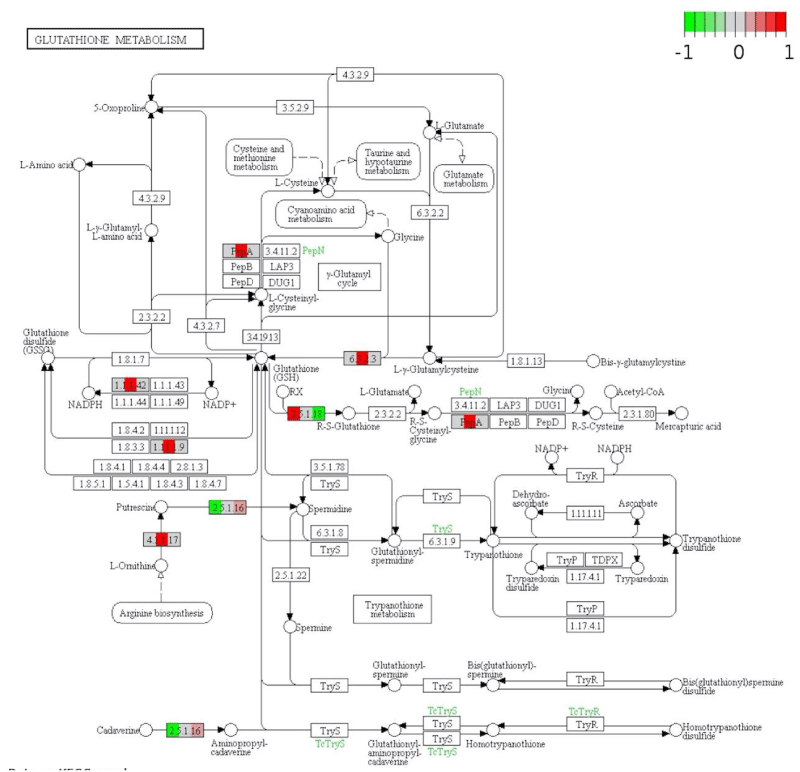
Many other major pathways were altered including glutathione metabolism (Figure 4), glycolysis (Figure S4), and TCA cycle (Figure S5). DNA replication (Figure S6) was also significantly altered. These pathways are essential to maintain an energy source for the cell. They play a central role in providing energy and precursors for various metabolic reactions of the cell [73]. Apart from these, purine (Figure S7) and pyrimidine biosynthesis (Figure S8) was altered in our analysis after exposure to AgNPs. Both these pathways play a major role in many cellular processes.
Materials and methods
Preparation of silver nanoparticles
AgNPs were synthesized using an aqueous extract of Artemisia absinthium [32]. The plant of A. absinthium was raised in a greenhouse and dried at room temperature. 1g of leaf tissue powder was boiled in 10 ml of deionized water for 5 min. The plant extract was cooled to room temperature and filtered through a 0.45 um filter. This aqueous extract and AgNO3 (2 mM) were then mixed in equal volumes. After 24 hr, the mixture was centrifuged at 14,000 × g for 10 min. This was repeated five times and the resulting AgNPs were used for in vitro analysis ([32]. The preparation and characteristics of these AgNPs has been reported previously [32].
Inhibition of P. parasiticain conditions in vitro
The data on the inhibitory potential of AgNPs on P. parasitica have been previously reported by Ali, et al. [32], where the study confirmed that AgNPs were highly potent in inhibiting mycelial growth, production and germination of zoospores, and germ tube elongation [32]. The AgNPs used in the study were synthesized previously by Ali, et al. [32] and the synthesis was confirmed by recording UV-vis spectra [32]. AgNPs were also characterized physically using transmission electron microscopy, dynamic light scattering and zeta potential [32].
Phytophthora parasitica culture and treatment with AgNPs.
parasitica isolate 13-724 (isolated from citrus) was cultured in 25 mL of 10% V8 medium in 250 mL flasks for 2 days at room temperature on a rotary shaker at 50 RPM. The mixture of 10% V8 juice, 3000 zoospores and AgNPs (25 µg ml−1) in 200 µl total reaction volume was assembled in flat-bottom microtiter plates. Controls were mock-treated with deionized water. Mycelia were then collected at four time points (0hr, 15 min, 1 hr and 15 hr) from two samples at each time point, and flash-frozen in liquid nitrogen. Total RNA was isolated from each sample at each time point time point 0 hr, 15 min, 1 hr, and 15 hr using the RNeasy Plant Mini Kit according to manufacturer’s protocols (Qiagen). The quality of RNA samples was checked by gel electrophoresis and Bioanalyser (Agilent 2100 Bioanalyser). RNA samples with RNA Integrity Numbers (RIN) greater than 8.5 were used for cDNA library construction for sequencing. Two replicates for each time point were used.
cDNA library construction, and RNA sequencing
Each mRNA sample was sequenced using the Illumina HiSeq 2000 platform at the Duke Genomics Sequencing facility according to Illumina protocols. Briefly, poly-A-tail containing mRNA was purified from the total RNA using poly(T)-Magnetic beads. Sequencing cDNA libraries were constructed according to the manufacturer’s protocols (Illumina, San Diego, CA). Each library was sequenced using the pair-end 125 cycles (2 x 125 bp) protocol. All RNA samples were indexed with different Illumina adapters and were run in one lane of the Illumina flow cell. Base-calling and quality values (Q) were calculated according to the Illumina data processing pipeline. Using these protocols, we obtained approximately 20 million high-quality reads per sample that provided substantial sequence data for in-depth differential gene expression analyses.
RNA-Seq data analyses
The raw reads were filtered through to eliminate the low-quality reads and maintain only the Phred score >20 depending on wrapper script Trim galore [74]. Trim galore was also used to remove the adapters and checked for quality of reads per sample using FastQC [75]. TopHat2 software [76] was used to map high quality reads to the reference genome P. parasiticaINRA 310.3( AGFV00000000.2) downloaded from the BROAD institute website (https://olive.broadinstitute.org/strains/phytophthora_parasitica_inra_310.2) ("(dataset) Broad Institute [35] Phytophthora parasitica INRA-310 Genome sequencing and assembly . National Center for Biotechnology Information,"). Tophat2 results were processed using SAMtools [77]. Generation of read count table for each gene was conducted using HTseq-count V0.6.1 [78] R package DESeq2 [79] was used to calculate differential gene expression. We used cutoff value of corrected p -values < 0.05 to select the deferentially expressed genes (Figure S9).
KEGG analysis
To visualize the involvement of the differentially expressed genes in biological pathways and look for the response of P. parasiticato AgNPs at 15 min, 1hr and 15hr, Kyoto Encyclopedia of Genes and Genomes (KEGG) ontology of differentially expressed genes was done by using KEGG Automatic Annotation Server (KAAS) and assigned KO terms for each group of genes were mapped on KEGG pathways using Pathview [80].
qRT-PCR analysis
Based on our exploratory RNA-Seq data, seven genes that were differentially expressed and had importance in many essential cellular processes were selected. The selected seven genes were subjected to quantitative real-time PCR using a previously published method [81]. Gene-specific primers (Table S6) were designed using NCBI primer blast tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). Ubiquitin-conjugating enzyme gene was used as a reference to calculate the relative expression values as it has been reported as a suitable internal control for the study of gene expression in P. parasitica [82]. Melting curve analysis was carried to verify the amplification of a specific single gene by each primer pair.
Conclusion
AgNPs have robust inhibitory effects on P. parasitica. AgNPs are known to inhibit multiple pathways, and can be utilized as an alternative way of controlling plant pathogens [83,84]. To the best of our knowledge, this is the first report where a comprehensive and in-depth analysis on the mode of action of AgNPs on P. parasitica using exploratory RNA-Seq analysis and qRT-PCR analysis was utilized to observe the changes in gene expression over time of 15 hrs. We found that AgNPs inhibit the respiratory chain enzymes, disrupt the ribosome biogenesis, glutathione metabolism, and purine and pyrimidine metabolism. Exposure to AgNPs affected genes involved in oxidative stress. All these changes significantly affect the viability of P. parasitica. The study emphasizes the potential of AgNPs as a new promising antimicrobial agent to control Phytophthora spp. However, further research is required to confirm the efficacy in field conditions.
Supplementary materials
Supplementary materials can be found in attachment and RNA-Seq data is deposited in NCBI.
Author contributions
ASES carried out the biological experiments. GSA designed the experiments. SB carried out the all analysis of expression data, qRT-PCR, Pathway analysis and drafted the manuscript. ZA participated in the qRT-PCR experiment and pathway analysis. All authors read and approved the final manuscript.
Funding
This work was supported by funds to GSA from the Florida Agriculture Experiment Station of the Institute of Food and Agricultural Sciences at the University of Florida. Additional support was provided by Fulbright Foundation to SB.
- Kroon LP, Brouwer H, de Cock AW, Govers F (2012) The genus Phytophthora anno 2012. Phytopathology 102: 348-364. Link: https://bit.ly/3lZTHOv
- Scholthof KBG (2007) The disease triangle: pathogens, the environment and society. Nat Rev Microbiol 5: 152-156. Link: https://bit.ly/3sgmv6t
- Fry W (2008) Phytophthora infestans: the plant (and R gene) destroyer. Mol Plant Pathol 9: 385-402. Link: https://bit.ly/3CIBOKh
- Tyler BM (2007) Phytophthora sojae: root rot pathogen of soybean and model oomycete. Mol Plant Pathol 8: 1-8. Link: https://bit.ly/3iMTaxE
- Cline ET, Farr DF, Rossman AY (2008) A Synopsis of Phytophthora with Accurate Scientific Names, Host Range, and Geographic Distribution. Plant Health Progress 9: 32. Link: https://bit.ly/2VOyUD8
- Prigigallo MI, Mosca S, Cacciola SO, Cooke DEL, Schena L (2015) Molecular analysis of Phytophthora diversity in nursery-grown ornamental and fruit plants. Plant Pathology 64: 1308-1319. Link: https://bit.ly/3seeAGM
- Beever RE, Waipara NW, Ramsfield TD, Dick MA, Horner IJ (2009) Kauri (Agathis australis) under threat from Phytophthora. Phytophthoras in forests and natural ecosystems 74: 74-85. Link: https://bit.ly/3AFtXLn
- Vannini A, Brown A, Brasier C, Vettraino AM (2009) The search for Phytophthora centres of origin: Phytophthora species in mountain ecosystems in Nepal. In Proceedings of Goheen EM, Frankel SJ (tech. coords). Proceedings of the Fourth Meeting of the International Union of Forest Research Organisations (IUFRO) Working Party S 54-55. Link: https://bit.ly/37HxIUi
- Meng Y, Zhang Q, Ding W, Shan W (2014) Phytophthora parasitica: a model oomycete plant pathogen. Mycology 5: 43-51. Link: https://bit.ly/2Xw4qq1
- Erwin DC, Ribeiro OK (1996) Phytophthora diseases worldwide; American Phytopathological Society (APS Press). Link: https://bit.ly/3xMqlpg
- Rossman AY, Palm ME (2006) Why are phytophthora and other oomycota not true fungi? Outlooks on Pest Management 17: 217-219. Link: https://bit.ly/3mgK12z
- Werner S, Steiner U, Becher R, Kortekamp A, Zyprian E, et al. (2002) Chitin synthesis during in planta growth and asexual propagation of the cellulosic oomycete and obligate biotrophic grapevine pathogen Plasmopara viticola. FEMS Microbiology Letters 208: 169-173. Link: Link: https://bit.ly/2VNzhNM
- Parra G, Ristaino JB (2001) Resistance to mefenoxam and metalaxyl among field isolates of Phytophthora capsici causing Phytophthora blight of bell pepper. Plant Dis 85: 1069-1075. Link: https://bit.ly/3m28nwB
- Lee TY, Mizubuti E, Fry WE (1999) Genetics of metalaxyl resistance inPhytophthora infestans. Fungal Genetics and Biology 26: 118-130. Link: https://bit.ly/3m3dixi
- Childers R, Danies G, Myers K, Fei Z, Small IM, et al. (2015) Acquired resistance to mefenoxam in sensitive isolates of Phytophthora infestans. Phytopathology 105: 342-349. Link: https://bit.ly/3fYO6UL
- Hu J, Hong C, Stromberg EL, Moorman GW (2010) Mefenoxam sensitivity in Phytophthora cinnamomi isolates. Plant Disease 94: 39-44. Link: https://bit.ly/2XieEtK .
- Meng Q, Cui X, Bi Y, Wang Q, Hao J, et al. (2011) Biological and genetic characterization of Phytophthora capsici mutants resistant to flumorph. Plant Pathology 60: 957-966. Link: https://bit.ly/3CPxTer
- Timmer LW, Graham JH, Zitko SE (1998) Metalaxyl-resistant isolates of Phytophthora nicotianae: occurrence, sensitivity, and competitive parasitic ability on citrus. Plant Disease 82: 254-261. Link: https://bit.ly/3CMbOxt
- Gisi U, Cohen Y (1996) Resistance to phenylamide fungicides: a case study with Phytophthora infestans involving mating type and race structure. Annu Rev Phytopathol 34: 549-572. Link: https://bit.ly/3CKVYTR
- Pal S, Tak YK, Song JM (2007) Does the antibacterial activity of silver nanoparticles depend on the shape of the nanoparticle? A study of the gram-negative bacterium Escherichia coli. Appl Environ Microbiol 73: 1712-1720. Link: https://bit.ly/3yOuJVQ
- Jo YK, Kim BH, Jung G (2009) Antifungal activity of silver ions and nanoparticles on phytopathogenic fungi. Plant Dis 93: 1037-1043. Link: https://bit.ly/3jTj2Hj
- Strayer A, Ocsoy I, Tan W, Jones J, Paret M (2016) Low concentrations of a silver-based nanocomposite to manage bacterial spot of tomato in the greenhouse. Plant Dis 100: 1460-1465. Link: https://bit.ly/3yN09fu
- Kim KJ, Sung WS, Suh BK, Moon SK, Choi JS, et al. (2009) Antifungal activity and mode of action of silver nano-particles on Candida albicans. Biometals 22: 235-242. Link: https://bit.ly/3xKV85P
- Clement JL, Jarrett PS (1994) Antibacterial silver. Met Based Drugs 1: 467-482. Link: https://bit.ly/3g0Iyt0
- Rai MK, Deshmukh SD, Ingle AP, Gade AK (2012) Silver nanoparticles: the powerful nanoweapon against multidrug‐resistant bacteria. J Appl Microbiol 112: 841-852. Link: https://bit.ly/37GVW0X
- Feng QL, Wu J, Chen G, Cui F, Kim T, et al. (2000) A mechanistic study of the antibacterial effect of silver ions on Escherichia coli and Staphylococcus aureus. J Biomed Mater Res 52: 662-668. Link: https://bit.ly/3yLopi3
- Yamanaka M, Hara K, Kudo J (2005) Bactericidal actions of a silver ion solution on Escherichia coli, studied by energy-filtering transmission electron microscopy and proteomic analysis. Appl Environ Microbiol 71: 7589-7593. Link: https://bit.ly/3xJPyR5
- Park HJ, Kim JY, Kim J, Lee JH, Hahn JS, et al. (2009) Silver-ion-mediated reactive oxygen species generation affecting bactericidal activity. Water Res 43: 1027-1032. Link: https://bit.ly/2VNzDUC
- Gordon O, Slenters TV, Brunetto PS, Villaruz AE, Sturdevant DE, et al. (2010) Silver coordination polymers for prevention of implant infection: thiol interaction, impact on respiratory chain enzymes, and hydroxyl radical induction. Antimicrob Agents Chemother 54: 4208-4218. Link: https://bit.ly/3iK0aew
- Liau S, Read D, Pugh W, Furr J, Russell A (1997) Interaction of silver nitrate with readily identifiable groups: relationship to the antibacterialaction of silver ions. Lett Appl Microbiol 25: 279-283. Link: https://bit.ly/2VMbjT3
- Holt KB, Bard AJ (2005) Interaction of silver (I) ions with the respiratory chain of Escherichia coli: an electrochemical and scanning electrochemical microscopy study of the antimicrobial mechanism of micromolar Ag+. Biochemistry 44: 13214-13223. Link: https://bit.ly/3yN0bE8
- Ali M, Kim B, Belfield KD, Norman D, Brennan M, et al. (2015) Inhibition of Phytophthora parasitica and P. capsici by silver nanoparticles synthesized using aqueous extract of Artemisia absinthium. Phytopathology 105: 1183-1190. Link: https://bit.ly/37IxhsY
- Sun D, Zhang W, Mou Z, Chen Y, Guo F, et al. (2017) Transcriptome Analysis Reveals Silver Nanoparticle-Decorated Quercetin Antibacterial Molecular Mechanism. ACS Applied Materials & Interfaces 9: 10047-10060. Link: https://bit.ly/3jSSaXW
- Ivask A, ElBadawy A, Kaweeteerawat C, Boren D, Fischer H, et al. (2014) Toxicity Mechanisms in Escherichia coli Vary for Silver Nanoparticles and Differ from Ionic Silver. ACS Nano 8: 374-386. Link: https://bit.ly/3COLsen
- (Dataset) Broad Institute (2018) Phytophthora parasitica INRA-310 Genome sequencing and assembly. National Center for Biotechnology Information.
- Singh N, Paknikar KM, Rajwade J (2019) RNA-sequencing reveals a multitude of effects of silver nanoparticles on Pseudomonas aeruginosa biofilms. Environmental Science: Nano 6: 1812-1828. Link: https://rsc.li/2VSJCrC
- Gullner G, Komives T, Király L, Schröder P (2018) Glutathione S-transferase enzymes in plant-pathogen interactions. Front Plant Sci 9: 1836. Link: https://bit.ly/3AV8H4L
- Shen M, Zhao DK, Qiao Q, Liu L, Wang JL, et al. (2015) Identification of glutathione S-transferase (GST) genes from a dark septate endophytic fungus (Exophiala pisciphila) and their expression patterns under varied metals stress. PloS One 10: e0123418. Link: https://bit.ly/3g2gd5s
- Hussain S, Hess K, Gearhart J, Geiss K, Schlager J (2005) In vitro toxicity of nanoparticles in BRL 3A rat liver cells. Toxicol In Vitro 19: 975-983. Link: https://bit.ly/3AFukFL
- Seo JH, Pyo S, Shin YK, Nam BG, Kang JW, et al. (2018) The Effect of Environmental Enrichment on Glutathione-Mediated Xenobiotic Metabolism and Antioxidation in Normal Adult Mice. Front Neurol 9: 425. Link: https://bit.ly/2VSJMPK
- Deponte M (2013) Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. Biochim Biophys Acta 1830: 3217-3266. Link: https://bit.ly/3xM6A19
- Halliwell B, Gutteridge JM (2015) Free radicals in biology and medicine; Oxford University Press, USA. Link: https://bit.ly/3m1EyfG
- Sellam A, Poupard P, Simoneau P (2006) Molecular cloning of AbGst1 encoding a glutathione transferase differentially expressed during exposure of Alternaria brassicicola to isothiocyanates. FEMS Microbiol Lett 258: 241-249. Link: https://bit.ly/3g1DzIL
- Liao S, Zhang Y, Pan X, Zhu F, Jiang C, et al. (2019) Antibacterial activity and mechanism of silver nanoparticles against multidrug-resistant Pseudomonas aeruginosa. Int J Nanomedicine 14: 1469-1487. Link: https://bit.ly/3ALvakD
- Hatem E, Berthonaud V, Dardalhon M, Lagniel G, Baudouin-Cornu P, et al. (2014) Glutathione is essential to preserve nuclear function and cell survival under oxidative stress. Free Radic Biol Med 1: S25- S26. Link: https://bit.ly/2VSmE3Y
- Ilyas S, Rehman A (2014) Oxidative stress, glutathione level and antioxidant response to heavy metals in multi-resistant pathogen, Candida tropicalis. Environ Monit Assess 187: 4115. https://bit.ly/2VT5vae
- Allocati N, Masulli M, Di Ilio C, Federici L (2018) Glutathione transferases: substrates, inihibitors and pro-drugs in cancer and neurodegenerative diseases. Oncogenesis 7: 8. Link: https://bit.ly/2VPXvXF
- Pigné S, Zykwinska A, Janod E, Cuenot S, Kerkoud M, et al. (2017) Flavoprotein supports cell wall properties in the necrotrophic fungus Alternaria brassicicola. Fungal Biology Biotechnology 4: 1. Link: https://bit.ly/3jS23Fs
- Forneris F, Binda C, Vanoni MA, Mattevi A, Battaglioli E (2005) Histone demethylation catalysed by LSD1 is a flavin‐dependent oxidative process. FEBS Lett 579: 2203-2207. Link: https://bit.ly/3yZKny5
- Gross E, Kastner DB, Kaiser CA, Fass D (2004) Structure of Ero1p, source of disulfide bonds for oxidative protein folding in the cell. Cell 117: 601-610. Link: https://bit.ly/3AE0OQQ
- Mattevi A (2006) To be or not to be an oxidase: challenging the oxygen reactivity of flavoenzymes. Trends Biochem Sci 31: 276-283. Link: https://bit.ly/3AMJadX
- Fraaije MW, Mattevi A (2000) Flavoenzymes: diverse catalysts with recurrent features. Trends Biochem Sci 25: 126-132. Link: https://bit.ly/2UhY503
- Hardilová Š, Havrdová M, Panáček A, Kvítek L, Zbořil R (2015) Hsp70 as an indicator of stress in the cells after contact with nanoparticles. In Proceedings of Journal of Physics: Conference Series 012023. Link: https://bit.ly/3ySHEXa
- Akerfelt M, Morimoto RI, Sistonen L (2010) Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol 11: 545-555. Link: https://bit.ly/3yO4W03
- Browning LM, Lee KJ, Cherukuri PK, Huang T, Songkiatisak P, et al. (2018) Single gold nanoparticle plasmonic spectroscopy for study of chemical-dependent efflux function of single ABC transporters of single live Bacillus subtilis cells. Analyst 143: 1599-1608. Link: https://rsc.li/3g2fMIE
- Garmory HS, Titball RW (2004) ATP-binding cassette transporters are targets for the development of antibacterial vaccines and therapies. Infect Immun 72: 6757-6763. Link: https://bit.ly/3seaSNA
- Kovács D, Szőke K, Igaz N, Spengler G, Molnár J, et al. (2016) Silver nanoparticles modulate ABC transporter activity and enhance chemotherapy in multidrug resistant cancer. Nanomedicine 12: 601-610. Link: https://bit.ly/37WkDXv
- Hayashi K, Schoonbeek HJ Sugiura H, De Waard MA (2001) Multidrug resistance in Botrytis cinerea associated with decreased accumulation of the azole fungicide oxpoconazole and increased transcription of the ABC transporter gene BcatrD. Pesticide Biochemistry and Physiology 70: 168-179. Link: https://bit.ly/3iKCyqj
- Lee YJ, Yamamoto K, Hamamoto H, Nakaune R, Hibi T (2005) A novel ABC transporter gene ABC2 involved in multidrug susceptibility but not pathogenicity in rice blast fungus, Magnaporthe grisea. Pesticide Biochemistry Physiology 81: 13-23. Link: https://bit.ly/3siMydb
- Nakaune R, Adachi K, Nawata O, Tomiyama M, Akutsu K, et al. (1998) A novel ATP-binding cassette transporter involved in multidrug resistance in the phytopathogenic fungusPenicillium digitatum. Appl Environ Microbiol 64: 3983-3988. Link: https://bit.ly/3AINLNY
- Chen L, Xie QW, Nathan C (1998) Alkyl Hydroperoxide Reductase Subunit C (AhpC) Protects Bacterial and Human Cells against Reactive Nitrogen Intermediates. Mol Cell 1: 795-805. Link: https://bit.ly/3xFTgv2
- Dayem AA, Kim B, Gurunathan S, Choi HY, Yang G, et al. (2014) Biologically synthesized silver nanoparticles induce neuronal differentiation of SH‐SY5Y cells via modulation of reactive oxygen species, phosphatases, and kinase signaling pathways. Biotechnol J 9: 934-943. Link: https://bit.ly/3sillHW
- Zhang XF, Shen W, Gurunathan S (2016) Silver nanoparticle-mediated cellular responses in various cell lines: an in vitro model. Int J Mol Sci 17: 1603. Link: https://bit.ly/3xL8KxO
- Manning G, Plowman GD, Hunter T, Sudarsanam S (2002) Evolution of protein kinase signaling from yeast to man. Trends Biochem Sci 27: 514-520. Link: https://bit.ly/3ABLqV4
- Cook A, Lowe E, Chrysina E, Skamnaki V, Oikonomakos N, et al. (2002) Structural studies on phospho-CDK2/cyclin A bound to nitrate, a transition state analogue: implications for the protein kinase mechanism. Biochemistry 41: 7301-7311. Link: https://bit.ly/3AFy8Hd
- Comfort KK, Maurer EI, Braydich-Stolle LK, Hussain SM (2011) Interference of silver, gold, and iron oxide nanoparticles on epidermal growth factor signal transduction in epithelial cells. ACS Nano 5: 10000-10008. Link: https://bit.ly/3jN3Vza
- Schönichen A, Webb BA, Jacobson MP, Barber DL (2013) Considering protonation as a posttranslational modification regulating protein structure and function. Annu Rev Biophys 42: 289-314. Link: https://bit.ly/3CJHQtY
- Hunter T (1995) Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell 80: 225-236. https://bit.ly/3AF5S7A
- Rauch J, Volinsky N, Romano D, Kolch W (2011) The secret life of kinases: functions beyond catalysis. Cell Commun Signal 9: 23. Link: https://bit.ly/3iHM3Gu
- Fromont-Racine M, Senger B, Saveanu C, Fasiolo F (2003) Ribosome assembly in eukaryotes. Gene 313: 17-42. Link: https://bit.ly/3xM7Q4n
- Chaillou T, Kirby TJ, McCarthy JJ (2014) Ribosome biogenesis: emerging evidence for a central role in the regulation of skeletal muscle mass. J Cell Physiol 229: 1584-1594. Link: https://bit.ly/3mgLh5N .
- Awad D, Prattes M, Kofler L, Rössler I, Loibl M, et al. (2019) Inhibiting eukaryotic ribosome biogenesis. BMC Biology 17: 46. Link: https://bit.ly/2VTz3VK
- Edirisinghe JN, Weisenhorn P, Conrad N, Xia F, Overbeek R, et al. (2016) Modeling central metabolism and energy biosynthesis across microbial life. BMC Genomics 17: 568. Link: https://bit.ly/3sgmTlo
- Krueger F (2015) Trim galore. A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files.
- Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom.
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, et al. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology 14: R36. Link: https://bit.ly/3xQbZ73
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078-2079. Link: https://bit.ly/2UmjQfe
- Anders S, Pyl PT, Huber W (2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166-169. Link: https://bit.ly/3seJI9l
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15: 550. Link: https://bit.ly/3AHR349
- Luo W, Pant G, Bhavnasi YK, Blanchard SG, Brouwer C (2017) Pathview Web: user friendly pathway visualization and data integration. Nucleic Acids Res 45: W501-W508. Link: https://bit.ly/3g2E9px
- El-Sayed AS, Yassin MA, Ali GS (2015) Transcriptional and proteomic profiling of Aspergillus flavipes in response to sulfur starvation. PloS one 10: e0144304. Link: https://bit.ly/3CQ1tAs
- Yan HZ, Liou RF (2006) Selection of internal control genes for real-time quantitative RT-PCR assays in the oomycete plant pathogen Phytophthora parasitica. Fungal Genet Biol 43: 430-438. Link: https://bit.ly/3iHMm46
- Kim SW, Jung JH, Lamsal K, Kim YS, Min JS, et al. (2012) Antifungal effects of silver nanoparticles (AgNPs) against various plant pathogenic fungi. Mycobiology 40: 53-58. Link: https://bit.ly/2Xt5nze
- Jung JH, Kim SW, Min JS, Kim YJ, Lamsal K, et al. (2010) The effect of nano-silver liquid against the white rot of the green onion caused by Sclerotium cepivorum. Mycobiology 38: 39-45. Link: https://bit.ly/3jOuCn4

Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.
 Save to Mendeley
Save to Mendeley
