Journal of Clinical Microbiology and Biochemical Technology
Bacteriophage Based Assays for Detection of Salmonella Organisms
Vimlesh Gupta and Hari Mohan Saxena*
Cite this as
Gupta V, Saxena HM (2016) Bacteriophage Based Assays for Detection of Salmonella Organisms. J Clin Microbiol Biochem Technol 2(1): 041-045. DOI: 10.17352/jcmbt.000015Salmonellosis is an important zoonotic disease but field applicable, simple, accurate, and cost effective diagnostics are lacking. We isolated a broad acting bacteriophage lytic to Salmonella Typhimurium, S. Pullorum, S. Gallinarum and S. Dublin and investigated its application in diagnosis employing MTT assay and nitrate test. On addition of MTT (3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) to live bacteria alone of isolates of Salmonella, its color changed from yellow to purple. However, after 4 hours of incubation of bacteria with bacteriophage, no color change was observed because of lysis of bacteria by the phage. After incubation with salmonellaphage, twelve out of forty clinical samples showed no colour change indicating lysis of Salmonella by phage, whereas samples without Salmonella showed colour change due to other bacteria. In nitrate test, live bacteria reduce nitrate to nitrite which can be detected by change in colour. No colour developed on prior incubation of Salmonella with the phage.
Introduction
Salmonellosis is a hyperendemic disease in India affecting humans, animals and poultry birds. Salmonellosis is caused by organisms of genus Salmonella. In poultry birds Salmonella enterica subsp. enterica serovars Gallinarum (biovars Gallinarum and Pullorum) and Enteritidis is the leading cause of morbidity and mortality and responsible for significant economic loss in poultry industry. Salmonella is a food borne enteric pathogen of great public health importance causing infection on consumption of eggs and meat from infected birds. Prevalence of Salmonella up to 14.7% in live poultry birds in India has been reported [1].
To prevent losses due to the disease, accurate diagnosis and correct and effective treatment in early stages is essential. Various advanced diagnostic tests e.g. ELISA, CFT, PCR etc. are available but these are not employed at field level because they are cumbersome, require very costly, sophisticated equipment and skilled personnel and can be performed in specialized labs only. Available ELISA and molecular assay based kits are very costly. There are limited diagnostic facilities under field conditions.
By employing bacteriophages along with suitable indicator systems, easy and accurate diagnosis of bacterial pathogens is possible in field conditions. The ability of bacteriophage to specifically infect, and lyse its host bacterium is to be exploited as a means of uniquely identifying target bacteria. Phage-based assays are particularly attractive since they are rapid, simple, and do not require the use of expensive equipment [2].
Materials and Methods
Collection of samples
A total of 102 samples comprising faecal swabs of live birds, spleen, caecum, liver and congested intestine of dead birds and uterine discharges, diarrhoeal faeces of cattle, buffalo and horse were collected in sterile containers from college clinics and different poultry farms, and clinics in and around Ludhiana. The samples were immediately transported on ice to the laboratory for further processing.
Tissue pieces of ceacum, liver and congested intestine were homogenized in pestle and mortar and fecal swabs of poultry and faeces of animals in 200 µl volume of RPV broth. The homogenate prepared by suspending the tissue in RPV broth, was inoculated on HEA/BGA plate for isolation. RPV medium was prepared by dissolving 4.92 g of Rappaport Vassiliadis medium (Himedia) in 100 ml of distilled water and autoclaving at 1210C, at 15 lbs pressure for 15 minutes. Hektoen Enteric Agar (HEA) medium was prepared by dissolving 7.66 g of hektoen enteric agar medium (Himedia) in 100 ml of distilled water and boiling it. After boiling, it was poured in petriplates and allowed to solidify.
The longitudinal surface of each tissue sample was incised with a sterile scalpel followed by introduction of a loop in the cut surface and the material brought with the loop was inoculated into RPV broth and on HEA/BGA plates. Fecal swabs of poultry, and feces and uterine fluid of animals were also inoculated into RPV broth and HEA/BGA plates. The inoculated samples were incubated at 370C for 24 hrs.
Identification and biochemical characterization of Salmonella
A colony was picked up from agar plate, smeared on microscopic glass slide and subjected to Gram’s staining and observed under a microscope. The following biochemical tests were performed as per the standard procedure [3], for confirmation of suspected colonies.
Catalase test: Two to three drops of 3% H2O2 were taken on a clean grease-free glass slide and single colony was mixed with the help of a wire loop. Immediate formation of gas bubbles was considered as a positive test.
Oxidase test: Commercially available standard oxidase discs (Himedia, Mumbai) were used to perform this test as per the method described by Quinn et al., [3]. Single colony was rubbed with the help of a sterile wire loop on the oxidase disc vigorously. Immediate development of blue/violet colour was considered as a positive test.
H2S production: The suspected colony was inoculated on TSI slant by stabbing the butt and by streaking the slant and incubated at 370Cfor 16 hours. In case of Salmonella, the tube showed changes in colour like alkaline slant (red), acidic butt (yellow), H2S production (black), and gas production (gas bubbles).
Motility Indole Lysine (MIL): One isolated colony was picked up and inoculated into motility indole lysine medium tube. The result was considered positive when the colour of medium changed from purple to turbid pink.
Citrate utilization test: The suspected colony was inoculated on Simmon citrate slant by streaking the slant and incubated at 370C for 12 hours. The test was considered positive after change in colour from green to blue.
Confirmation of Salmonella isolates by polymerase chain reaction (PCR)
The suspected isolates were confirmed by PCR method by using Salmonella genus specific oligonucleotide primers of 25 base pairs [4].
Extraction of genomic DNA: The genomic DNA of Salmonella isolates was extracted as per the method of Wilson [5]. Briefly bacteria from broth culture were lysed and proteins were digested with proteinase K. Cell wall debris, polysaccharides and remaining proteins were removed by selective precipitation with CTAB. The high molecular weight DNA was recovered from resulting supernatant by isopropanol precipitation.
PCR: The PCR assay was carried out by using Salmonella genus specific oligonucleotide primers of 25 base pairs. These were got synthesized from Promega as per the sequence given in Table 1.
Isolation of salmonellaphage
24 Sewage samples were collected from Veterinary Clinics of GADVASU, Ludhiana and from herd suspected to be Salmonella affected (based on clinical history and symptoms) in 50 ml wide mouth samplers and were transported on ice to the laboratory for further processing.
Double strength NZCYM broth was used for primary inoculation of sewage sample. For this 1.15g NZCYM broth was dissolved in 25ml distilled water and sterilized by autoclaving at 1210C at 15 lbs for 15 minutes. For preparing agar for pouring over plates, 2.3g of NZCYM broth and 0.75g bacteriological agar (Himedia, Mumbai) were dissolved in 100ml of distilled water, boiled and then dispensed in 5 ml screw capped vials. The medium was sterilized by autoclaving at 1210C at 15 lbs for 15 minutes. NZCYM agar plates were used for Salmonellaphage isolation. For these 2.3g NZCYM broth and 2g bacteriological agar were dissolved in 100 ml of distilled water. The medium was then autoclaved at 1210C at 15 lbs for 15 minutes. After cooling the medium was poured into sterile petriplates.
Salmonella Enteritidis strain (procured from IVRI Izatnagar) was used for isolation of Salmonellaphage from sewage samples.
20 ml of sewage and 5 ml overnight grown Salmonella Enteritidis culture were added into 25 ml of double strength NZCYM broth and then incubated at 370C for up to 24 hrs. The processed sample was inoculated by agar overlay technique as per McDuff et al. [6], for bacteriophage isolation. 1.5ml of processed sample was transferred into a microcentrifuge tube (2 ml, Eppendorf) and centrifuged at 10000 rpm for 10 minutes. The supernatant was collected and filtered through a pre-sterilized 0.22µm PVDF filter (Axiva). Later on 600 µl of filtrate and 200 µl overnight grown broth culture of S. Enteritidis were mixed in 0.75% NZCYM agar (maintained at 450C) and was spread over 1.5% NZCYM plates. The plates were allowed to solidify at room temperature and incubated at 370C aerobically for 24 hrs.
Plates having plaque formation were preserved and the plaques were confirmed for the presence of phage by secondary streaking.
200 µl overnight grown broth culture of S. Enteritidis was mixed in 3 ml of 0.75% NZCYM agar (maintained at 45oC) and was spread over 1.5% NZCYM plates. The plates were allowed to solidify at room temperature. The plaques suspected for Salmonellaphage were picked up by using platinum wire inoculation loop and streaked firstly as horizontal lines across the plates, later on as vertical lines dissecting the horizontal lines across the plate at 900. The plates were then incubated at 370C aerobically for 24 hrs. The clearing along the streak line indicated the presence of bacteriophage. For further use phage was eluted from the plates into sterile SM buffer. Sterile SM buffer (5 ml) was poured over suspected bacteriophage plates showing clearance around streak lines. The plaques along line were disturbed using the platinum loop and the plates were kept in incubator at 370C up to 8 hrs. The elutant was collected and centrifuged to discard any agar particle. Later on supernatant was filtered through 0.22µm PVDF filter and then preserved at 40C.
Heterogeneity test of phage
The lytic activity of Salmonellaphage was tested against Salmonella Typhimurium, S. Pullorum, S. Gallinarum, and S. Dublin (procured from IVRI, Izatnagar) and field isolates as well as several bacteria of heterologous species viz. Staphylococcus aureus, Brucella species, Escherichia coli, and Pasteurella multocida.
200 µl overnight grown broth culture of test bacteria was added to 3 ml of 0.75% NZCYM agar (maintained at 450C) and mixed well and poured over 1.5% NZCYM agar plates. The plates were allowed to solidify at room temperature. The plaques suspected for Salmonellaphage were picked by using platinum wire inoculation loop and streaked as secondary streaking. The plates were then incubated at 370C aerobically for 24 hrs.
Phage based diagnostic assays
Various assays were attempted employing the lytic phage to determine if they can be used for diagnostic purpose. These assays included: MTT assay, Nitrate reductase detection assay and Catalase detection assay.
Standard Salmonella cultures (S. Enteritidis) were obtained from IVRI (Izatnagar). Bacteriophages against Salmonella isolated in our lab were used for the development of the test protocol. The standard cultures of Salmonella were inoculated on BSM and TSB/RPV broth respectively and incubated at 370C overnight.
Phage based MTT assay
100 µl of overnight grown culture of Salmonella spp. was added to the microtitre plate and to this equal volume of Salmonellaphage was added. Control wells included Salmonella, phage, and broth alone only. The plate was incubated at 370C for 61/2/4 hours respectively.
After incubation 20 µl of MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) / (Thiazolyl blue tetrazolium bromide) (Himedia) was added in each well at a concentration of 5 mg/ml (prepared in PBS), and change in colour was observed within 5 minutes [7].
Phage based nitrate reductase detection assay
100 µl of overnight grown culture of Salmonella spp. was placed in the microtitre plate. To this equal volume of salmonellaphage was added. Control wells included Salmonella, phage, and broth alone only. 20 µl nitrate broth was added in each well and incubated at 370C for 61/2/4 hours respectively. After incubation, 20 µl each of reagent A and reagent B were added.
Phage based catalase detection assay
100 µl of overnight grown culture of Salmonella was placed in the microtitre plate, and equal volume of Salmonellaphage was added. Control wells included Salmonella, phage, and broth alone only. 10 µl H2O2 was added in each well and incubated at 370C for 4 hours.
After incubation, 100 µl of TMB/H2O2 was added in each well and any change in colour was observed within 5 minutes.
Results and Discussion
Collection of samples
A total of 102 samples comprising of fecal swabs of live birds, spleen, ceacum, liver and congested intestine of dead birds and uterine discharges and diarrhoeal faeces of cattle, buffalo and horse were collected.
Isolation of Salmonella organisms from samples
Out of a total of 102 clinical samples, 9 were positive on isolation (7 from uterine fluid and 2 from emu birds) (Table 2).
Identification and biochemical characterization of Salmonella isolates
Identification of Salmonella isolates was done on the basis of cultural and staining characterstics and biochemical parameters. Light pink colonies on BGA and green colonies with black centre on HEA were observed. The organisms appeared as gram negative rods.
The colonies were subjected to biochemical tests like H2S production, catalase, oxidase, motility indole lysine (MIL) and citrate utilization test. All the 9 isolates were found to be negative for oxidase and positive for catalase, motility indole lysine (MIL), and citrate utilization test and all were lysed by Salmonellaphage isolated in present study.
Confirmation of isolates by PCR
The genomic DNA of field isolates of Salmonella and the standard culture strain of Salmonella Enteritidis were extracted by using standard bacterial DNA extraction method described by Wilson [5]. The OD260/280 value for isolated DNA was ~1.8-1.9 for all samples indicating the purity of DNA.
The extracted DNA from Salmonella isolates and Salmonella Enteritidis when subjected to PCR by using Salmonella genus specific oligonucleotide primers of 25 base pairs revealed the desired amplicons of 496 bp as per Cohen et al. [4].
Isolation of Salmonellaphage
Out of 24 sewage samples processed, Salmonellaphage could be isolated from 1 sample. Upon streaking the plaques on Salmonella lawn, clear zones along the streak lines were obtained (Figure 1). At a dilution of 10-6, the phage concentration was 4.17 × 109 plaques per ml which was countable. The isolated Salmonellaphage lysed Salmonella Typhimurium, S. Pullorum, S. Gallinarum and S. Dublin (procured from IVRI, Izatnagar) and all the 9 Salmonella field isolates but did not lose any of the heterologous species viz. Staphylococcus aureus, Brucella species, Escherichia coli, and Pasteurella multocida.
Phage based diagnostic assays
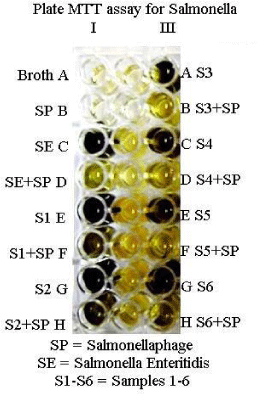
Phage based MTT assay: In the present study, attempts were made to develop a phage based diagnostic assay employing MTT (3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide), harnessing the ability of live bacteria to produce the reductase enzyme which causes a change in the colour of the redox dye (MTT) from yellow to purple in contrast to no colour change reaction after incubation of the bacterial suspension with the specific bacteriophage owing to lysis. All the standard isolates of Salmonella indicated colour change from yellow to purple upon addition of MTT after 4 hours of incubation of live bacteria alone. However, in case of wells containing the specific bacteriophage-bacteria suspension, no colour change was observed after 4 hours of incubation because of the specific lysis of bacteria by the phage.
Based on the above concept, 40 samples for salmonellosis suspected birds from a poultry farm were screened for salmonella. It was observed that 12 samples showed no colour change after 4 hours of incubation with Salmonellaphage. However colour change was observed for other samples (Figures 2,3). This indicates that Salmonella was present in those 12 positive samples. Montoro et al. [7], reported that in MTT assay change in colour from yellow to violet indicated the bacterial growth and that no colour change means death of bacteria. In the present study, the colour change was correlated with the death of bacteria due to the specific phage. A bacteriophage based MTT assay for Brucella has already been reported by us [8].
Phage based nitrate reductase detection assay
This test is based on the ability of live bacteria to reduce nitrate to nitrite due to the release of nitrate reductase by bacteria. The presence of nitrite can be easily detected by nitrate reagents which produce change in colour. No colour developed when salmonella were incubated with the phage. Montoro et al. [7], reported that a change in colour indicated the bacterial growth and no colour change means death of the bacteria. Although the assay worked well on pure culture, it could not be adapted to clinical samples due to contaminating bacteria.
Phage based catalase detection assay
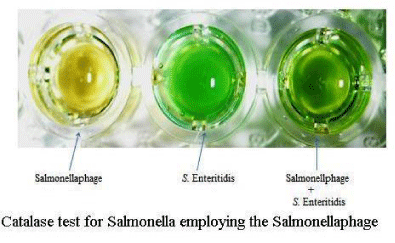
Colour changed to green in case of live Salmonella but in case of bacteria incubated with the phage, there was no colour change (Figure 4). However the assay worked only with pure culture of Salmonella but not with clinical samples due to contaminating bacteria. The assays may possibly be adapted to clinical samples after decontaminating them for other bacteria by using selective antibiotics.
The use of bacteriophage in assays for detecting bacteria was first reported over half a century ago when an assay to detect Salmonella using the phage Felix 01 was described by Cherry et al., [9]. Phage based diagnosis is easy because phages can be maintained at room temperature. McDuff et al. [6], observed that after lyophilisation, phage retained its activity during storage for at least 20 months at 40C. Phage is stable in broth at pH values from 6 to 8 for 24 hours at 370C.
Favrin et al. [10], developed an assay that utilized the normal infection cycle of bacteriophage SJ2 for detection of Salmonella enterica serovar Enteritidis in broth. The end point of the assay could be determined by using either fluorescence or optical density measurements. The detection limit of the assay in broth was less than 104 CFU/ml, and the assay could be performed in 4 to 5 h.
Chen and Griffiths [11], constructed recombinant bacteriophages specific for Salmonella spp. and containing either lux AB or the entire lux operon. By employing a 6 hour preincubation step of this reporter phage and Salmonella, as few as 10 Salmonella cells per ml in the original sample could be detected.
Summary and Conclusions
We investigated the possibility of application of a new lytic bacteriophage in the MTT assay, Nitrate reductase assay and Catalase assay for detection of Salmonella. In the MTT assay, twelve out of forty clinical samples showed no colour change after 4 hours of incubation with Salmonellaphage whereas colour change was observed for other samples indicating that Salmonella was present in those 12 positive samples. The Nitrate reductase assay and Catalase assay worked well only on pure culture, and could not be adapted to clinical samples due to contaminating bacteria.
- Murugkar HV, Rahman H, Kumar A, Bhattacharyya D (2005) Isolation, phage typing and antibiogram of Salmonella from man & animals in Northeastern India. Indian J Med Res 122: 237-242. Link: https://goo.gl/G7DI6f
- Gupta V, Saxena HM (2016) Phage based diagnosis of bacterial infections. J Clin Pathol Microbes 1: 1- 4. Link: https://goo.gl/B31nhK
- Quinn PJ, Carter ME, Markey B, Carter GR (1999) Clinical Veterinary Microbiology Mosby International Limited. Edinburgh 261-67. Link: https://goo.gl/7rXveM
- Cohen, N.D., Wallis, D.E., Neibergs, H.L., Hargis, B.M. (1995) Detection of Salmonella enteritidis in equine faeces using the polymerase chain reaction and genus-specific oligonucleotide primers. Journal of Veterinary Diagnostic Investigation 7: 219-222. Link: https://goo.gl/w2fybf
- Wilson K (1987) Preparation of genomic DNA from bacteria. In: Ausubal FM, Brent R, Kirston Rl, Moore DD, Seidman JG, Smith JA and Struhl K (Eds), Current Protocols in Molucular Biology 1. John Wiley and Sons, New York 241-242. Link: https://goo.gl/21TSuP
- McDuff CR, Jones LM, Wilson JB (1961) Characteristics of brucellaphages. J Bacteriol 83: 324-329. Link: https://goo.gl/uiNF3T
- Montoro E, Lemus D, Echemendia M, Martin A, Portaels F, et al. (2005) Comparative evaluation of the nitrate reduction assay, the MTT test, and the resazurin microtitre assay for drug susceptibility testing of clinical isolates of Mycobacterium tuberculosis. J Antimicrob Chemother 55: 500-505. Link: https://goo.gl/N67NhU
- Gupta V, Saxena HM (2013) Application of bacteriophage in the diagnosis of Brucellosis. Veterinary Practitioner 14(2) Suppl.1: 533-535. Link: https://goo.gl/weVlm3
- Cherry WB, Davis BR, Edwards PR, Hogan RB (1954) A simple procedure for the identification of the genus Salmonella by means of a specific bacteriophage. J Lab Clin Med 44: 51-55. Link: https://goo.gl/IPmX2E
- Favrin SJ, Jassim SA, Griffiths MW (2001) Development and optimization of a novel Immunomagnetic Separation Bacteriophage Assay for detection of Salmonella enterica Serovar Enteritidis in broth. Applied and Environmental Microbiology 67: 217-224. Link: https://goo.gl/gOw1fs
- Chen J, Griffiths MW (1996) Luminescent Salmonella strains as real time reporters of growth and recovery from sublethal injury in food. Int J Food Microbiol 31: 27-43. Link: https://goo.gl/85r04d

Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
