Journal of Clinical Microbiology and Biochemical Technology
Halotolerant Co-Cultured Bacterial Strains used in Degradation of Tannin Isolated from Tannery Industry Contaminated Sites
Veena Gayathri Krishnaswamy1*, Raiza waizd1 and Gopika2
2Department of Biochemistry, University of Madras, India
Cite this as
Veena gayathri K, Raiza W, Gopika (2016) Halotolerant Co-Cultured Bacterial Strains used in Degradation of Tannin Isolated from Tannery Industry Contaminated Sites. J Clin Microbiol Biochem Technol 2(1): 009-014. DOI: 10.17352/jcmbt.000009The residual tannins discharged from tanning units of the leather industry have an adverse effect on living organism and causes serious environmental pollution. Biological degradation is an important mechanism of organic chemical removal in natural systems owing to its environmental compatibility. But in extreme environment like high salinity of tannery effluent, the efficiency of degradation by microorganisms would be exceptionally low. Therefore halotolerant microbes would play a significant role in biodegradation of tannins. This study aims at isolating and identifying strains of halotolerant co-cultured bacteria able to degrade tannin from soil samples contaminated by tannery industry, in order to use such bacteria for the bioremediation of contaminated sites. The bacterial isolates have been characterized by biochemically and molecularly through 16S rRNA sequencing, and identified as Bacillus amyloliquefaciens and Bacillus polyfermenticus, this latter being able to degrade 92% of tannin (200 mg/L) compared to B. amyloliquefaciens. The variables (salinity, carbon and nitrogen sources) affecting the process of tannin degradation have been optimized.
Introduction
Tannery industry is one of the oldest industries in the world. With the growth of population, the increasing requirement of leather and its products led to the establishment of large commercial tanneries [1]. Tanneries are typically characterized as pollution intensive industrial complexes which generate varying high-strength wastewaters. Tanning industry is one of the highest toxic intensity per unit of output [2]. Tannery effluent is among one of the hazardous pollutants of industry. Major problems are due to wastewater containing heavy metals, toxic chemicals, chloride, lime with high dissolved and suspended salts and other pollutants [3]. Tanneries generate wastewater in the range of 30-35 Lkg-1 skin/hide processed with variable pH and high concentrations of suspended solids, BOD, COD, tannins including chromium [4].
Tannins are water soluble polyphenolic secondary metabolites of higher plants which are either galloyl esters or their derivatives. It is the second most abundant plant phenolic compound present after lignin [5]. At industrial scale tannin is used in leather production and wood staining [6]. After lignins, they are the second most abundant group of plant phenolics. Their tanning property is due to their capacity to combine with proteins. However, they can also complex with other polymers such as alkaloids, cellulose, and pectins [7].
Tannins have long been considered toxic to the microorganisms and this activity is mainly due to enzyme inhibition and substrates deprivation, action on ion deprivation [8]. Nevertheless, some fungi, yeasts and bacteria are quite resistant to tannins and can also degrade them [9,10]. Amongst bacterial species with the ability to degrade these polyphenolics are Citrobacter spp., Streptococci spp. and Cornybacterium spp [11].
Many conventional processes were carried out to treat tannery wastewater, such as physical treatment, chemical treatment and biological treatment [12-16]. Among these, physical and chemical methods are considered very expensive in terms of energy and reagents consumption and generation of excessive sludge [17]. The main advantages of biological treatment methods are they have low capital and operating costs compared to alternatives such as chemical-oxidation processes. There will be true destruction of organics, versus mere phase separation and oxidation of a wide variety of organic compounds, removal of reduced inorganic compounds, such a sulphides and ammonia. At the end of the treatment there is reduction of aquatic toxicity [18]. One drawback of biological treatment is, it is time consuming and involves the biological nature of the microbes for the degradation.
Normally existing neutrophilic bacteria can grow only with normal pH and in salt content less than 3.5 %, increase in the salinity will inhibit the growth of these microbes. Hence, it is very difficult for the neutrophilic bacteria to remove tannin using neutrophiles. In such places there is a need of Archae group of bacteria which contains extremophilic organisms which could be employed to remove the tannins. Halophiles are group of extremophilic bacteria which can survive and tolerate salinity. Veena Gayathri and Vasudevan [19] worked on the degradation of tannin at high concentrations by a bacterial consortium isolated from tannery effluent soil. The tannin concentrations were up to 500 mg/L by the bacterial consortium. The degradation of tannin was relatively toxic at 500mg/L than other concentrations where the removal efficiency lowered to 75%. Halophilic bacteria are a group of archae bacteria which can survive in high salt content and also has the capability to degrade xenobiotic compounds. Tannery effluents are industrial effluents with high salt content and higher chemical oxygen demand. There is a need for halophilic microbes which could easily grow in those recalcitrant compounds in the presence of salt. Halophilic microbes which could grow well in the presence of NaCl have been employed for the biodegradation which has proven effective as they could grow well and degrade the pollutant [20]. Halophiles are microbes which grow optimally in media containing salt between 3% and 15% NaCl [21].
Hence in the present study, tannin degrading halotolerant bacterial co-cultures were isolated from the tannery effluent contaminated soil and their growth conditions were optimized for efficient degradation of tannins. The bacterial strains present in the co-cultures were biochemically characterized and identified by 16S rRNA sequencing.
Materials and Methods
Enrichment of Tannin degrading Bacterial consortium
Soil samples collected from contaminated site of tannery industry situated in Pallavaram, Tamil nadu, India where raw hides are processed for isolation of tannin degrading bacterial strains. One gram of soil was added to 100ml mineral salts medium containing tannin (0.1%) and incubated at 150C under shaking (120 rpm). The bacterial co-cultures were enriched in mineral salts medium (g/L) of NaCl 10.0 to 150.0, KH2PO4 0.25, NH4Cl 1.0, Na2BO7 2.0, FeCl3 0.0125, CaCl2 0.06 and MgCl2 0.05. The medium was supplemented with a specified amount of added NaCl and 10mg of yeast extract, adjusted to pH-7 [22]. The medium was autoclaved, cooled to room temperature and was amended with tannin through a sterile filter (0.45micron) in 250ml Erlenmeyer flasks. The chemicals and reagents used in the study were analytical grade. The tannin containing agar plates were inoculated with 0.1ml suspension from these flasks. The isolated bacterial colonies were transferred to the tannin containing mineral salts medium. The pure culture was maintained on tannin containing mineral salts agar slants at 40C.
Growth of the bacterial strains on tannin was studied by determining the colony forming units (CFU) per ml on nutrient agar medium containing tannin. The mineral medium was substituted with different concentrations of NaCl (50g/L to 70g/L), while the isolated co-cultured bacterial strains was able to show an optimum degradation at 50mg/L. At 25 h time interval, the culture was centrifuged at 10,000rpm for 15 minutes to remove the cells. The culture supernatant was collected and used for the estimation of residual tannin.
Degradation of tannin
For the degradation study, mineral salts medium containing tannin (50mg/L to 200mg/L) was inoculated with the bacterial co-cultures. Experimental set up used for the degradation of tannin were (i) medium + tannin + bacterial co-cultures (ii) medium + tannin (iii) medium + bacterial cocultures, with (ii) and (iii) serving as controls. The bacterial co-cultures were added to the medium at concentrations of 104-105 cfu/ml. The culture, in duplicate was incubated at 370C with shaking at 150rpm. The cell suspensions were clarified by centrifugation at (10,000rpm for 15min at 60C). Biodegradation of tannin by bacterial co-cultures were observed till the end of their growth phase for 72h [21].
Effect of salt, carbon and nitrogen sources
Influence of salt on the degradation of tannin as studied by changing the salt concentrations to 3% and 7% NaCl. To study the effect of different carbon sources (0.5%), additional carbon sources like glucose, fructose, sucrose was added in the medium. The growth medium containing yeast extract was replaced with alternate nitrogen sources such as ammonium sulphate, potassium nitrate, ammonium nitrate, beef extract to study the effect of different nitrogen sources.
Identification of tannin degrading bacterial strains
The individual bacterial strains were separated from the bacterial co-cultures, which was used in the degradation of the tannin. The bacterial strains present in the consortium were examined using conventional biochemical tests initially. Molecular identification of the bacterial strains was done by 16S rRNA sequencing.
Biochemical characterization
The bacterial strains present in the consortium were isolated and grown separately as pure cultures. Initially, Gram staining and motility test were performed after which biochemical characterization was done for different tests (Motility, Triple sugar iron agar, Catalase, Urease, Indole production, Methyl red, Voges Proskauer, Citrate utilization) using 24h old culture. After 24h incubation at 370C, the colour change observed was accounted for positive/negative result. Genus level identification of the unknown bacterial strains were accomplished by using Bergey’s Manual of Systematic Biology [23] to ascertain the existence of variable biochemical test results for each strain.
16S rRNA partial gene sequencing
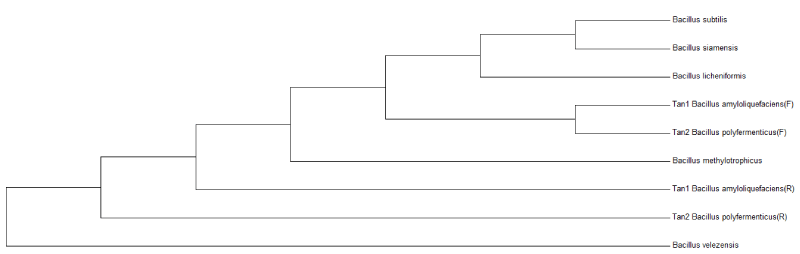
Chromosomal DNA was isolated from pure strains of the consortium by the standard phenol/chloroform extraction method (22). The 1.5 kb partial sequence of 16S rRNA gene was amplified by the chromosomal DNA using poly-merase chain reaction (PCR) with universal Eubacteria-specific primers 16F27 (5_-CCA GAG TTT GAT CMT GGC TCA G-3_) and 16R1525XP (5_-TTCTGCAGT CTA GAA GGA GGT GWT CCAGCC-3_). The PCR amplification conditions were: initial denaturation at 94C for 2 min, followed by 35 cycles of denaturation at95C for 1 min, annealing at 55C for 1 min, extension at 72C for 1 min, and a final extension at 72C for 10 min; and finally sequencing was performed on an ABI310-automatedDNA sequencer using Big Dye terminator kit (Applied Biosystems 3730xl DNA Analyzer). The amplified 16S rRNA gene (PCR products) from these isolates was directly sequenced after purification by precipitation with polyethylene glycol and NaCl. The primers used to obtain the complete sequence of 16S rRNA gene of the isolates were the same as used for the PCR amplification (16F27N and 16R1525XP). Sequence analysis was performed using Chromas Prosequence analysis software. The phylogenetic tree was constructed by MEGA6 software.
Results and Discussion
Isolation and characterization of Tannin degrading bacterial consortium
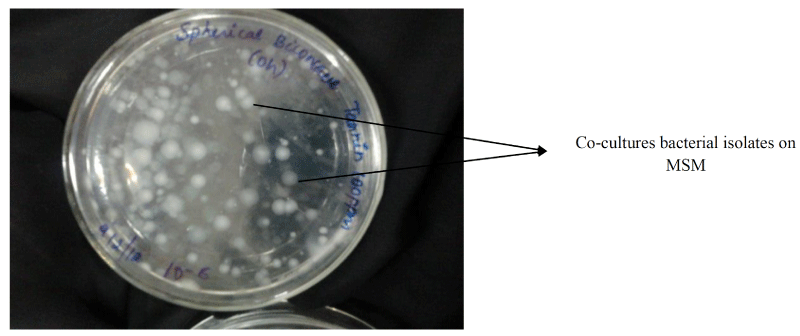
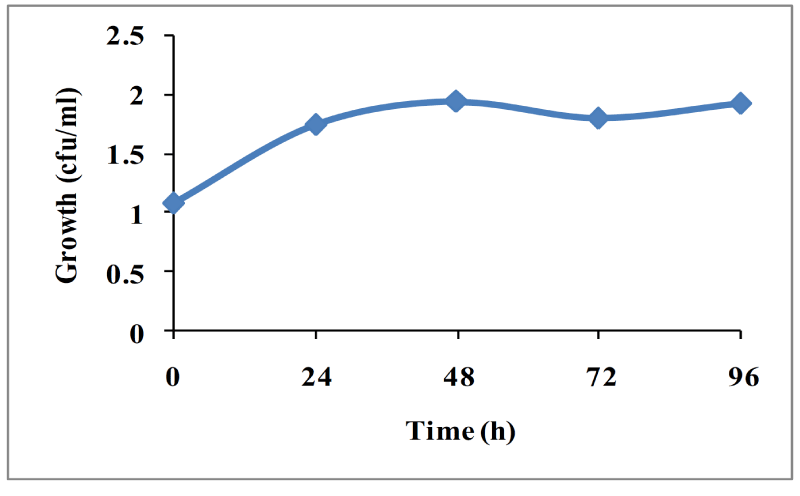
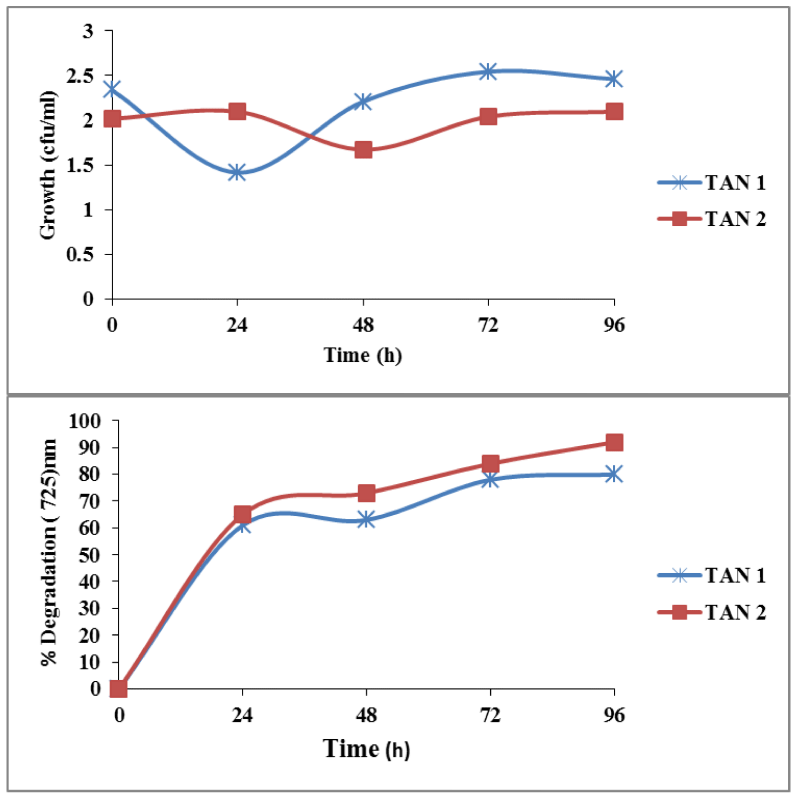
Bacterial strains isolated from the soil samples were screened on tannin containing agar plates. During isolation period, several bacterial strains co-existed in the consortium which could degrade tannin (50mg/L) under saline conditions (Figure 1). After successive transfer during enrichment period, two bacterial strain were isolated (TAN1, TAN2) which could survive and degrade tannin as a sole source of carbon and energy. The Figure 2 shows that there was an increase in viable cell count till 24h and by the end of 72 h, there was decrease in viable cell count indicating that bacterial co-cultures could not survive after 96h. Hence all the further experiments, the growth and degradation of tannin were monitored up to 96 h. The bacterial co-cultures were gram positive. The biochemical characteristics of two bacterial strains showed that they belong to the phyla Firmicutes. These groups of bacteria are commonly present in the contaminant soil, water and wastewater. Table 1 summarizes the results on the biochemical tests of the isolated bacterial strains. The two bacterial strains showed round colony morphology in the mineral salts agar plates. TAN 1 showed translucent raised colony with irregular edges. TAN 2 was small transparent flat colony with smooth edges and was big yellow mucoid colony with smooth edges. The both the bacterial isolates belongs to firmicutes in the order of Bacillales and the genus Bacillus based on the Bergys manual of classification. Through 16S rRNA sequencing results, the strains were identified as B. amyloliquefaciens strain (TAN 1), B. polyfermenticus strain (TAN 2). Phylogenetic tree analysis of the co-cultures strains was constructed from 16s r DNA and the evolutionary history was inferred using the Neighbor-joining method. The evolutionary distances were computed using the Kimura 2-parameter method. The phylo tree with the sum of branch length = 0.18268090 is shown in the Figure 3.
Degradation of Tannin at different concentrations by bacterial co-cultures
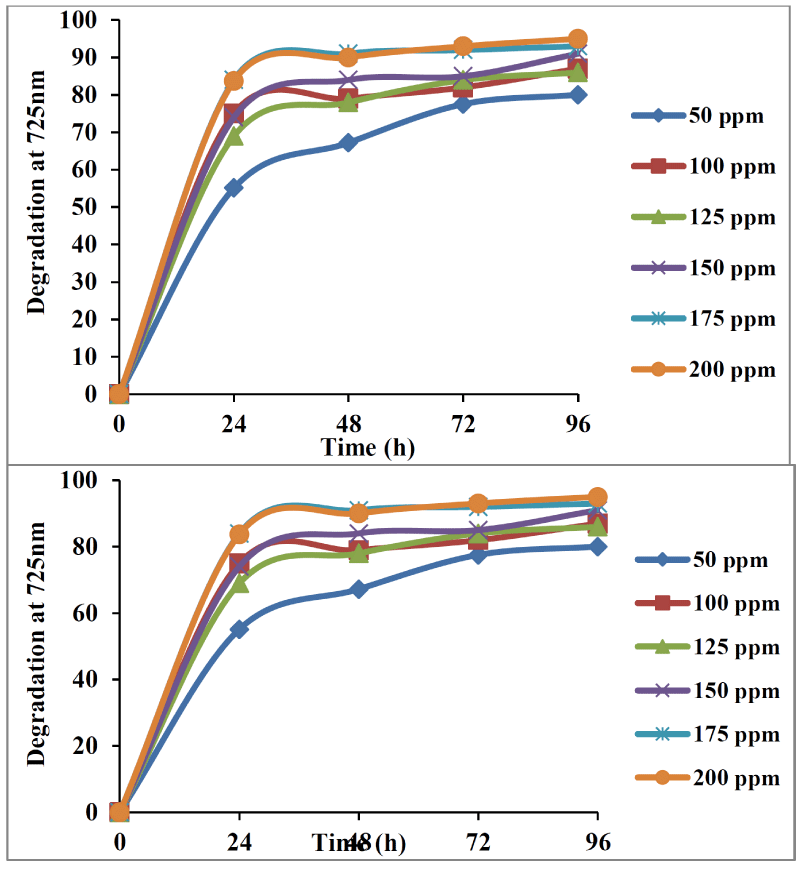
The ability of co-culture bacterial strains which could utilize tannin as a sole carbon source and energy was studied at 50mg/L to 200 mg/L (50mg/L, 100mg/L, 125mg/L, 150mg/L, 175mg/L, 200mg/L) of tannin. Tannin at 50mg/L of concentration showed minimum degradation i.e. up to 55% by the end of 24h and final degradation being 80 % by the end of 96 h. Maximum growth was observed in 50 mg/L concentration of tannin during 48 to 72 h of incubation. This was followed by 100 mg/L of tannin concentration showing degradation up to 87% by the end of 96 h. The batch experiments were repeated for other concentrations of tannin. When the concentration was increased to 200mg/L, the degradation was maximum of about 95% by 96 h and there was gradual decrease in the growth of organism. Therefore bacterial cocultures used in the present study could utilize tannin up to a concentration of 200mg/L with showed the optimum degradation (Figure 4).
Degradation of different concentrations of tannin by the individual bacterial strains
Bacterial strains present in the co-cultures were studied individually for the degradation of tannin at the optimum concentration 200 mg/L of tannin. TAN2 showed maximum degradation of tannin (200 mg/L) up to 92% by the end of 96h, when compared to TAN1 showing degradation up to 80% respectively. Viable cell in case of TAN2 increased from 0h 218x10-6 CFU/ml to 24h and there was subsequent decrease in cell count to 52x10-6 CFU/ml by the end of 72h (Figure 5).
Effect of Salt, Carbon and Nitrogen sources on tannin degradation
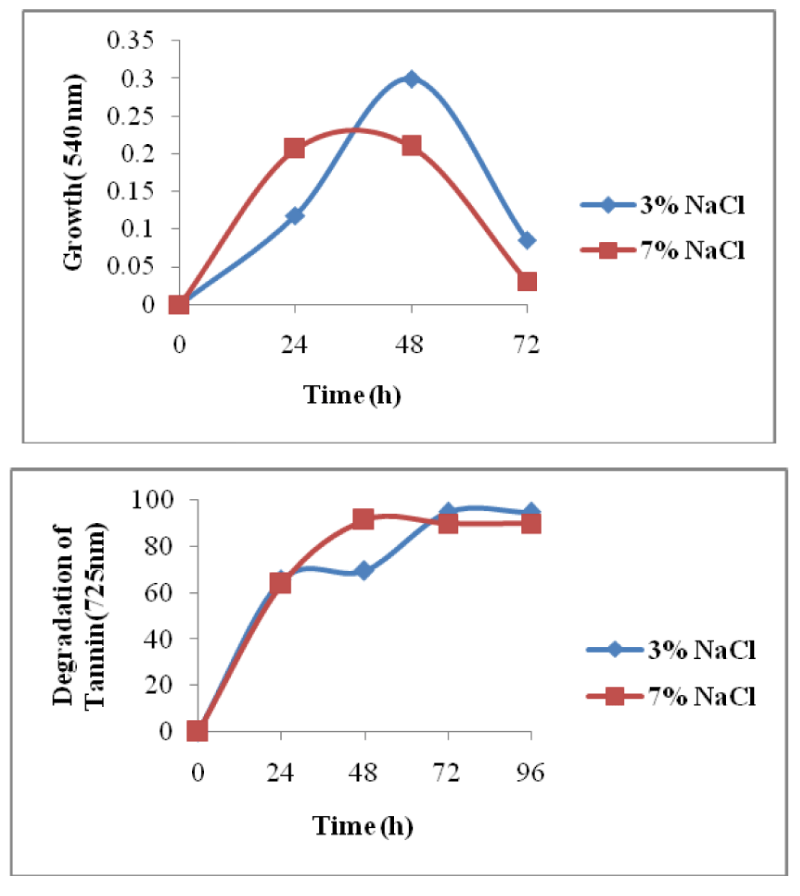
Salinity plays an important role in the metabolism of halophilic bacteria. The bacterial co-cultures were studied at different NaCl concentrations from 3% to 10%, while the cultures were able to survive only upto 7%. Hence the degradation experiments were carried out at 3% & 7% concentration at 200mg/L of tannin. Maximum degradation of 95% was observed at 3%NaCl and there was also simultaneous increase in the growth showing maximum growth (Figure 6) in 48 h of incubation. At 7% NaCl degradation reached up to 90%.
Figure 7 showed that, almost all the carbon sources were able to enhance the degradation of tannin. A great advantage for this degradation ability was particularly observed in the presence of glucose as carbon source showing degradation up to 91.4%. This was followed by sucrose and lactose having 90 and 84.7% of degradation respectively. Maximum growth was observed in glucose supplemented media followed by lactose and sucrose. Fructose didn’t play an important role in improving the degradation as only 69% of degradation was observed. Even the growth of organism in fructose supplemented media was less when compared to other carbon sources used.
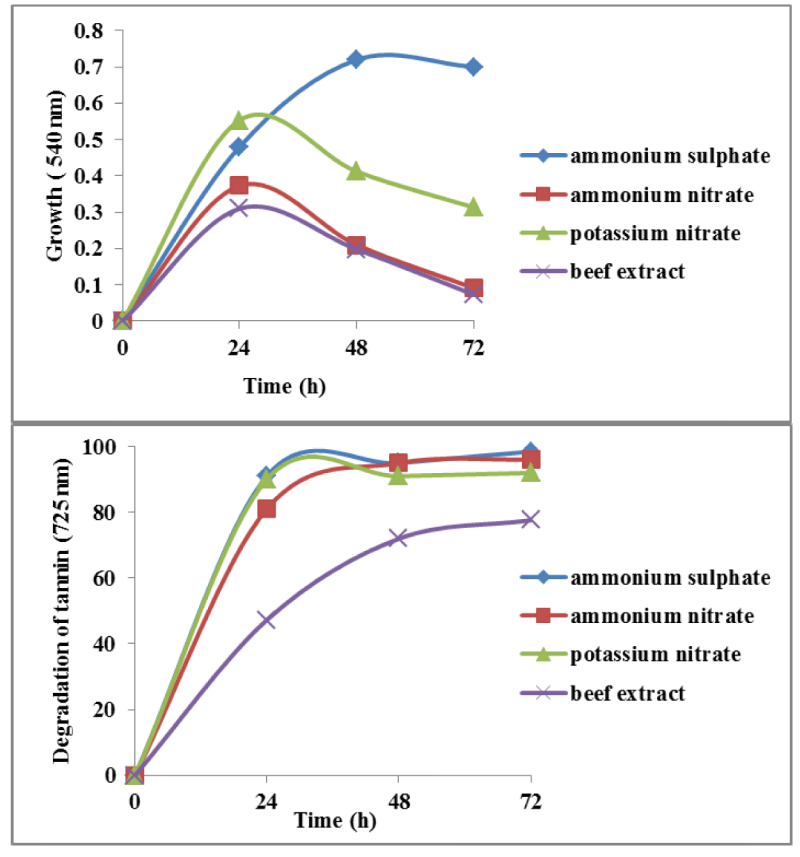
Figure 8 shows that ammonium sulphate, ammonium nitrate and potassium nitrate played an important role in enhancing the growth and degradation of tannin, ammonium sulphate being the maximum showing degradation up to 98% by the end of 96 h. This was followed by ammonium nitrate and potassium nitrate showing 96 and 92% of degradation respectively by the end of 96 h.
Maximum growth was observed in the ammonium sulphate supplemented media. Beef extract didn’t play an important role in enhancing the degradation as only 77.6% of degradation of tannin was observed.
Discussion
The residual tannins discharged from tanning units of the leather industry have adverse effects on living organisms and causing serious pollution to the environment. Tannins have long been considered toxic to microorganisms and this activity is mainly due to enzyme inhibition and substrate deprivation, action on membranes and metal ion deprivation [24]. Many authors reported microorganisms which are able to degrade tannins and to use them as carbon and energy source [25]. In the present study, the isolated bacterial consortium consisting of two bacterial strains, which could survive and degrade tannin up to 200 mg/L at 7% NaCl concentration was identified as B. amyloliquefaciens strain (TAN 1), B. fermenticus strain (TAN 2) through biochemical characterization and 16S rRNA sequencing.
Karthikeyan et al. [26], reported that the strain Klebsiella pneumonia could degrade tannin at 1% (w/v) and said that the optimum temperature for the tannase production is 35 °C ± 2. The degradation study of Tannin by a novel tannase enzyme present in some Lactobacillus plantarum strains by Jimenez et al. (2014), provided the advantage for the initial degradation of complex tannins present in plant environments [27].
Optimizing the condition for the biodegradation is very important for the efficient degradation of the compound. Parameters like pH, temperature, addition of carbon and nitrogen source and salinity in case of halophilic organism play an important role in optimization of the condition.
Salinity plays an important role in the metabolism of halophilic bacteria. At 3% NaCl concentration maximum degradation was obtained up to 95%. At 5% NaCl concentration degradation decreased 91%, followed by 7% at 200 mg/L tannin. Nitiema et al. [28], isolated Streptococcus species from anaerobic Shea cake digester, reported that optimum NaCl concentration for growth and degradation of tannic acid at 1 g/L concentration was 5 g/L and range for growth was 0 – 20 g/L. Carbon and nitrogen source also play an important role in the optimizing the conditions for degradation. The two strains TAN 1, TAN 2 could grow their be and degrade tannin up to 91.4% in the media supplemented with 0.5% of glucose. Umesh et al. [29], reported that glucose in the concentration of 1% in Bushnell and Hans’s medium could enhance the growth and degradation of tannin by Klebsialla spp.
Among nitrogen sources used in the present study for the optimization, ammonium sulphate could enhance the growth of the organism and the degradation reached maximum upto 98.5% followed by ammonium nitrate and potassium nitrate showing degradation upto 96 and 92%. Umesh et al. [29], used ammonium sulphate, peptone, ammonium persulphate, beef extract, malt extract, and tryptone to enhance the degradation of tannin and he reported that all nitrogen sources produced improvements in tannin biodegradation and ammonium sulfate in particular produced maximum tannin degradation upto 98%. Karthikeyan et al. [24], who worked with Klebsiella pneumonia, reported that among the nitrogen source tested in their study, inclusion ammonium nitrate resulted in the highest enzyme production which plays an important role in degradation of tannin.
Conclusions
In the present study, the halophilic bacterial consortium isolated from the polluted soil of tannery industry resulted in an effective biodegradation process in under saline conditions. The knowledge of substrate concentrations, effect of carbon, nitrogen and salt concentration on growth profiles, is needed in order to select strains with the required properties for biodegradation of tannins in saline environments. Hence the nutritive parameters were optimized for the efficient degradation of tannins under saline conditions. Such promising the halotolerant bacterial co-cultures could be used in the treatment of tannery effluent or to bioremediate tannery effluent contaminated soil.
The authors would like to thank the Management of Stella Maris College for providing opportunity to perform research and financial assistance. We would like to thank N. Madhana Priya Department of Bioinformatics for the phylogenetic analysis.
- Ahn DH, Chang WS, Yoon TI (1999) Dyestuff wastewater treatment using chemical oxidation, physical adsorption and fixed bed biofilm process. Process Biochem 34:429-439. Link: https://goo.gl/0hk4C
- Khan SR, Kawaja MA, Khan AM, Ghani H, Kazmi S (1999) Environmental impacts and mitigation costs associated with cloth and leather exports from Pakistan. A Report on Trade and Sustainable development Submitted by Sustainable Development Policy, Institute and IUCNP to IISD Canada for the IISD/IUCN/IDRC Project on Building Capacity for Trade and Sustainable Development in Developing Countries, Islamabad. Link: https://goo.gl/3s8TDO
- Uberoi NK (2003) Environmental Management. Excel Books Publisher, New Delhi: 269.
- Nandy, T, Kalu SN, Shastry S, Manivel W, Deshpande CV (1999) Waste-water management in cluster of tanneries in Tamilnadu through implementation of common treatment plants. J Sci Ind Res, 58: 475-516. Link: https://goo.gl/JzcpiZ
- Haslam E (1979) In Biochemistry of plant phenolics plenum press New York, 475-523.
- Van Soest PJ (1994) Nutritional Ecology of the Ruminant. Cornell University Press, Ithaca, New York. Link: https://goo.gl/Pecwa7
- Deschamps AM (1989) Microbial Degradation of Tannins and Related Compounds. In: Lewis NG, NG Paice NG (Eds.), Plant Cell Wall Polymers Biogenesis and Biodegradation. American Chemical Society, Washington, DC, 559-566. Link: https://goo.gl/RJsKPK
- Reed JD (1995) Nutritional toxicology of tannins and related polyphenols in forage legumes. J Anim Sci 73: 1516-1528. Link: https://goo.gl/wId4mo
- Scalbert A (1991) Antimicrobial properties of tannins. Phytochemistry 30: 3875-3883. Link: https://goo.gl/0NNGK7
- Mingshu L, Kai H, Qiang, Dongying J (2006) Biodegradation of Gallotannins and Ellagitannins. Journal of Basic Microbiology 46: 68-84. Link: https://goo.gl/Dsrb2M
- Ahn DH, Chung YC, Yoo YJ, Pak DW, Chang WS (1996) Improved treatment of tannery wastewater using Zoogloea ranigera and its extracellular polymer in an activated sludge process. Biotechnol Let 18: 917-922. Link: https://goo.gl/Miskrg
- Vijayaraghvan K, Murthy DVH (1997) Effect of toxic substances in anaerobic treatment of tannery wastewater, Bioprocess Biosys Eng 16: 151-155. Link: https://goo.gl/dfN7Uu
- Wiemann M, Schenk H, Hegemann W (1998) Anaerobic treatment of tannery wastewater with simultaneous sulphide elimination. Water Research 32: 774-780. Link: https://goo.gl/VbJrr
- Di Iaconi C, Lopez A, Ramadori R, Passino R (2003) Tannery wastewater treatment by sequencing batch biofilm reactor. Environ Sci Technol 37: 3199-3205. Link: https://goo.gl/nuVa80
- Farabegoli, G, Carucci, M, Majone, Rolle E (2004) Biological treatment of tannery wastewater in the presence of chromium, Journal of Environmental Management 71: 345-349. Link: https://goo.gl/sxQYMn
- Churchley JH (1994) Removal of sewage effluent- the use of a full scale ozone plant. Water Science and Technology 30: 275-284. https://goo.gl/skjBzU
- Stern SR, Azpyrkowicz, Rodighiro I (2003) Aerobic treatment of textile dyeing wastewater. Water Sci Technol 47: 55-59. Link: https://goo.gl/aBxcrG
- Bhat T, Singh B, Sharma O (1998) Microbial degradation of tannins-a current perspective. Biodegradation 9: 343-357. Link: https://goo.gl/O0La8q
- Veena Gayathri K, Vasudevan N (2010) Degradation of tannin at high concentrations by a bacterial consortium isolated from tannery effluent soil”. Biotechnology –an Indian Journal 4: 14-18. Link: https://goo.gl/ghpckQ
- Sivaprakasam S, Mahadevan S, Sekar S, Rajakumar S (2008) Biological treatment of tannery wastewater by using salt-tolerant bacterial strains. Microbial Cell Fact 7: 15-15. Link: https://goo.gl/OnyD2U
- Alva V, Peyton BM (2003) Phenol and catechol biodegradation by haloalkaliphile Halomonas campisalis: influence on pH and salinity. Environment Sci Technol 37: 4307-4402. Link: https://goo.gl/bcztBR
- Makkar HPS, Singh B, Karma DN (1994) Biodegradation of tannins in oak (Quercus incana) leaves by Sporotrichum pulverulentum. Lett Appl Microbio 18: 39-41. Link: https://goo.gl/qubXl8
- Bergey’s Manual of Systematic Bacteriology (2005) 2nd edn, In: Brenner DJ, Kreig NR, Stanley JT Springer, New York. Link: https://goo.gl/5OjmWw
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor NY.
- Reed JD (1995) Nutritional toxicology of tannins and related polyphenols in forage legumes. Journal of Animal science 73: 1516-1528. Link: https://goo.gl/wId4mo
- Karthikeyan S, Gurunathan J (2011) Production and partial purification of extra cellular tannase by Klebsiella pneumonia MTCC 7162 isolated from tannery effluent. African J Biotech 10: 364-1374. Link: https://goo.gl/4eLCr5
- Jimenez N, Esteban-Torres, Mancheno JM, De Las Rivas B, Munoz R (2014) Tannin degradation by a novel tannase enzyme present in some lactobacillus plantarum strains. Appl Environ Microbiol 80: 2991-2997. Link: https://goo.gl/A58YPS
- Nitiema LW, Dianou D, Simpore J, Karou SD, Savadogo PW, et al. (2010) Isolation of tannic acid degrading Streptococcus sp. from an anaerobic shea cake digester. Pak J Biological Science 13: 146-150. Link: https://goo.gl/BcQUXF
- Umesh J, Sudhir K, Nilesh T, Manohar P (2011) Degradation of tannic acid by cold adapted Klebsiella sp NACASAI and phytotoxicity assessment of tannic acid and its degradation product. Environ. Sci Pollut Res 18: 1129-1138. Link: https://goo.gl/a0FAoM

Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
