Journal of Clinical Research and Ophthalmology
A Laboratory Study on the Molecular Basis of Primary Congenital Glaucoma
Grand Chikezie Ihesiulor1, Forbes Manson2 and Udo Ahanna Ubani1*
2Division of Genomic Studies, University of Manchester, England
Cite this as
Ihesiulor GC, Manson F, Ubani UA (2018) A Laboratory Study on the Molecular Basis of Primary Congenital Glaucoma. J Clin Res Ophthalmol 5(1): 014-022. DOI: 10.17352/2455-1414.000049Purpose: To detect pathogenic mutations in cytochrome P450 family1 subfamily B polypeptide1 (CYP1B1) gene in nineteen sporadic Primary congenital glaucoma (PCG) cases and to identify patients lacking CYP1B1 mutations.
Methods: CYP1B1 exon 2 and the coding part of exon 3 of 15 participants were amplified by Polymerase chain reaction and amplicons were sequenced by Sanger sequencing. Sequencing data was analyzed to identify the gene mutations or Single Nucleotide Polymorphisms SNPs.
Results: Four previously reported PCG-associated CYP1B1 mutations (c.1159G>A; p.E387K, c.230T>C; p.L77P, c.1103G>A; p.R368H and c.1568G>A; p.R523K) were found in four patients out of the 15 fully ‘sequenced’ patients. Also, ten previously reported Single Nucleotide Polymorphisms and two novel noncoding variants were identified.
Conclusion: The relatively low percentage of PCG patients having CYP1B1 mutations (4/15=26.6%) demonstrates that other known and unknown genes may contribute to PCG pathogenesis. Lack of CYP1B1 gene mutations in some patients stresses the need to identify other responsible candidates
Introduction
Primary congenital glaucoma (PCG) is a classical form of infant buphthalmos and a predominant type of congenital glaucoma [1,2]. Although, buphthalmos has been reported from the time of Hippocrates (460-377 BC), it was associated with high intraocular pressure (IOP) and vision loss in children in the middle of the eighteenth century [1,3,4]. In PCG, isolated maldevelopment of ocular drainage structures increases resistance to aqueous outflow [3,5,6]. This leads to increased IOP, optic nerve atrophy and blindness if prompt and proper diagnosis and management is not provided [4,7,8].
Infants with PCG are affected differently. However, presentation of epiphora, blepharospasm, photophobia and swollen or cloudy cornea due to high IOP, is the norm [7].
Familial, like sporadic, PCG is an autosomal recessive disease [9]. It is highly prevalent in the consanguineous and inbred Gypsy subpopulation of Slovakia. This was reported to be due to genetic drift or ‘founder effect’ [8,10]. In non Gypsy populations, PCG may be inherited in a multifactorial fashion [11].
Although the genetic component of PCG is debatable, 4 chromosomal loci has been mapped including, GLAUCOMA 3, PRIMARY CONGENITAL, A (GLC3A on 2p22.2 locus; OMIM 231300), GLC3B (1p36.2-p36.1; OMIM 600975), GLC3C (14q24.2; OMIM 601771) and GLC3D on 14q24.3 locus (OMIM 613086).
Recently, PCG phenotype was associated with mutations in cytochrome P450 family 1subfamily B polypeptide 1 (CYP1B1) gene on the 2p21 locus, gene symbol GLC3A (OMIM #231300) and Latent transforming growth factor beta-binding protein 2 (LTBP2) gene (GLC3D; OMIM 602091) [8]. The genes associated with GLC3B and GLC3C are not yet known [12].
CYP1B1 gene analysis is the focus of this study. CYP1B1gene (Ensembl transcript ESNT00000260630) is a three-exon gene in which only exons 2 and part of 3 are translated [9]. It encodes a 543-amino-acid ‘drug metabolizing’ protein or enzyme involved in early anterior chamber angle development [13]. Mutations in CYP1B1 have also been implicated in other anterior segment dysgenesis (ASD) syndromes [14]. The molecular role of CYP1B1 gene in the pathophysiology of PCG is not fully understood [8]. This study identifies unrelated PCG subjects that have or lack CYP1B1 mutations by screening the coding exons of the CYP1B1gene using direct sequencing procedures. Patients having no disease-causing CYP1B1 alleles were noted for exome sequencing to find out other genetic factors involved in PCG pathogenesis [9].
Materials and Methods
Participants
Informed consent was obtained from the parents or responsible guardians of the subjects. The probands and their immediate families were questioned on their past medical history and examined at the hospital. Reviewing the eye charts from their respective eye physicians helped confirm their visual function. The inclusion criteria were intraocular pressure ≥21mmHg in the first years of life, the presence of cornea oedema, scar, or Haab striae or an ocular history consistent with infantile glaucoma. The presence of enlarged globe in their teens or adulthood suggests a history of congenital glaucoma in the first few years of life. Other clinical signs were examined using portable slit lamp and direct ophthalmoscope.
Congenital glaucoma CYP1B1 mutations (Table 1)
The study did not report the individual visual acuity, visual field and intraocular pressure of the patients.
DNA quantification and quality assessment
In order to determine the concentration and quality of extracted DNA (patient samples), the Nanodrop 8000 (Thermo Scientific) was used to obtain the absorbance of the DNA samples at 260nm; A260=1 was noted as the equivalent to 50ng/ml for double stranded DNA. The Nanodrop machine was ‘blanked’ initially by adding of nuclease-free water1µl (dH2O) onto the tip at the base of the spectrophotometer. The concentration and 260/280 values were noted (not shown). A 260/280 absorbance ratio of ≥ 1.8 was considered a better amount of purity for DNA samples. Samples with concentration of more than 100ng/µl were diluted properly with dH2O to make 100ng/µl concentration.
The DNA samples (in 100ng/ul concentrations) were spun down for 3-5 secs using Minispin at a speed of 2000rpm to prevent DNA from being shattered.
Designing primers
In order to design primers of the CYP1B1 gene (DNA) target, the following databases were used; ensembl; a massive database/genome browser, primer3plus and reverse complement.
Human CYP1B1 (ENST00000260630) gene sequence was obtained from ensembl. Oligonucleotide sequences were synthesized by Eurofins MWG Operon.
Primers were designed to include exons and a minimum of 50 base pairs (bp) of introns for desired sequence to be completely amplified.
Polymerase chain reaction (PCR) optimization
The best conditions for PCR were determined using gradient PCR. Primers were hydrated with a given volume of dH2O (Table 2) [15,16] (Acharya et al. 2006), and vortexed for 5-7 secs for uniform mixing of primers. Concentration varied between 5 and10 pmol/µl. For a 10 pmol/µl concentration, 90µl of dH2O was added to 10µl of primers. The ‘master mix’ tubes contained; Reddy mix (NEB or Promega): 12.5µl x 14 =175µl; Control DNA: 1.5 µl x 14 = 21µl; dH2O: 6.5µl x 14 = 91 µl; (Dimethyl sulfoxide) DMSO: 2.5 µl x 14 = 35µl; Total 23µl x 14 = 322µl
The different proportions were multiplied by 14 (see above) to account for pipette error. DMSO was added (for F1R1 primers only) to prevent formation of spurious secondary structures. The Promega (GoTaq® Green Master mix) contains bacterially derived Taq polymerase 2 X Green GoTaq® Reaction buffer (pH 8.5), 400µM dATP, 400µM dGTP, 400µM dCTP, 400µM dTTP and 3mM MgCl2. The New England Biolabs (NEB) (OneTaq quick load, 2X MM w/std buffer M0486S) contains dNTPs, MgCl2, buffer components and stabilizers. The NEB was used due to less smearing compared to Promega. PCR was performed under appropriate conditions and according to manufacturer's guideline.
Agarose gel electrophoresis
1% (w/v) agarose gel concentration was used throughout the study.
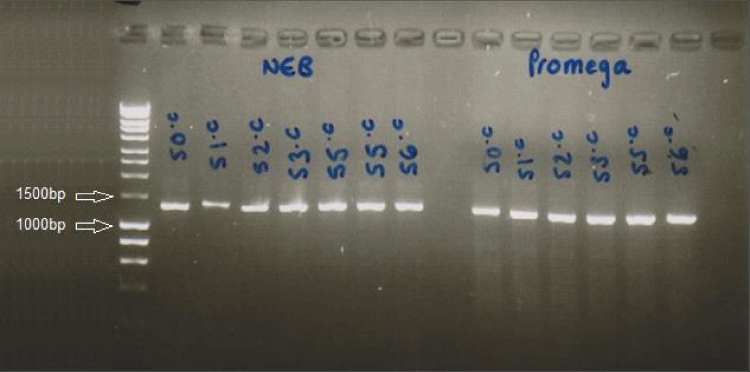
Gels were made with 1g of agarose powder (molecular grade) introduced into 100ml of 1x TAE (980ml of dH2O + 20ml of TAE buffer) buffer in a conical flask. After swirling gently, agarose-TAE mixture was heated for 2mins until the liquid was clear and the powder had dissolved completely. After cooling it slightly, 10µl SafeView (NBS Biologicals) was added. The SafeView dye is safer (less harmful) and stains DNA effectively to allow its visualization within the Agarose gel. After swirling gently and pouring the gel into the mold, a comb was placed in the second groove of the mold. After 20-30 minutes of gel setting, the combs were removed and the tray and gel were introduced into the electrophoresis tank containing sufficient 1 x TAE buffer. 5µl appropriately sized DNA ladder (hyperladder1 or 100bp #3231S) was loaded with pipette into the first well. 5µl of each amplicon was then loaded into the remaining wells carefully. Gels were electrophoresed at 100V voltage, 400Amps for 20 - 60 mins.
After electrophoresis, the gel was visualized to show the size (length) of the amplicon. This was carried out by UV-transillumination (Fluorchem imager) and an integrated camera was used to capture the images (Figure 1).
PCR amplification
In order to efficiently amplify the desired target DNA, a standard protocol was used in all reactions. Standard 25µl PCR reactions contained 12.5µl reddy mix (NEB), 2.5µl DMSO, 7µl dH2O, 1µl forward primer, 1µl reverse primer (Invitrogen) and 1µl DNA template.
After PCR optimization, the right Tm for Primer F1R1 and F3R3 were determined as 55oc and 56oc respectively. *Taq DNA polymerase extends the target strand at a rate of approximately 1kb per minute. A negative control (containing 1µl of dH2O instead of DNA) was also incorporated while setting up PCR reactions.
PCR products purification/cleaning
PCR plate reactions were purified on Agencourt AMPure XP magnetic beads (Beckman Coulter, High Wycombe) using a Beckman Coulter Biomek NX robot (Beckman Coulter, High Wycombe). Individual PCR amplicons were purified using Montage spin columns (Millipore Ltd, Watford) according to the manufacturer’s instruction.
Capillary- electrophoresis based chain-termination sanger sequencing
Taq polymerase enzyme, deoxynucleotide triphosphatases (dNTPs) and fluorescently-labelled, chain-terminating dideoxynucleotidetriphosphatase (ddNTP) were used in standard proportions. Purification of the reaction is carried out to eliminate the unincorporated ddNTPs [17].
In this study, the 10 µl sequencing reaction (master mix) was set up skirted well (PCr) plates with BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). it contained 0.25 µl BigDye v3.1, 1.875 µl Sequencing buffer, 5.375µl dH2O±0.5µl, 1-1.5µl of the AMPure purified amplicons, 1µl each of forward or reverse primer (in different rows). After mounting on the thermocycler (Verti 96 well, AB), the following conditions for sequencing were used; 96oc for 2mins, 96oc for 10sec, 55oc for 20 secs, 60oc for 4 mins, 8oc for 10mins, 4oc hold for infinity.
Purification of sequencing reactions
In order the remove the unincorporated dye terminator, Agencourt CleanSEQ paramagnetic bead solution (Beckman Coulter) and Beckman Coulter Biomek NX robotics were used. Multichannel pipette was used to introduce 5µl of CleanSEQ beads into skirted well (PCR tubes containing the sequencing product). The purification procedure was performed according to manufacturer’s guideline.
Sanger sequencing analysis
Two ABI files (sequence traces) for each patient, were created and named forward and reverse primer (for example JH5 EXON 2F.abi and JH5 EXON 2R.abi respectively). These files, and the reference sequence (CYP1B1) obtained from ensemble (ENST00000260630), were uploaded into the GeneScreen program.
The variants (boxes/cells) were edited and the chromatogram displayed was observed. Confirmed variants occurred in both forward and reverse reads. Strong peaks in the chromatogram were only regarded as real sequence changes. After analysis, samples with poor results were repeated. 5-10 nucleotides including variant identified were manually copied and matched on the cDNA sequence from ensembl. The number (cDNA position) was noted. Reference SNP (rs) number and other information such as chromosomal location were obtained using ensemble or Exome Variant Server (EVS).
The staden program helps to efficiently analyse sequences and identify variants/ mutations in DNA sequence. The Staden Pregap4.1.4b1 package was used to align the trace files to the desired reference database. First, the CYP1B1 (ENST00000260630) gene sequence obtained from UCSC genome browser was saved in a text file and uploaded to the Staden Pregap4.1.4b1 package. Then the Staden Gap package helped in analysing the contig (trace sequence) and comparing it with a known normal control (cell line DNA).
Results
CYP1B1 mutational analysis
Exons 2 and 3 were (Polymerase chain reaction) PCr-amplified and sequenced for each patient as described in the Materials and Methods. Sequence files were read with either the GeneScreen or Staden programs. One patient DNA (sample AK1) failed to give readable sequence (in both exons) despite multiple attempts. 15 out of 19 patients had both exons fully sequenced (Figure 1).
In tables 1,3 non-synonymous mutations were identified in five probands. In one patient only a single mutation was found meaning that the pick-up rate for CYP1B1 mutation being pathogenic in this panel of 15 ‘fully sequenced’ patients was 4/15 (26.67 %). All 5 mutations had been previously reported. Four (75%) of these mutations were found in exon 3 while one (25%) was identified in exon2. Exon 3 was fully sequenced in all 19 patients, while exon 2 was only fully sequenced in 15.
Out of the five probands that had two mutations, three were homozygous for a CYP1B1 mutation and one was compound heterozygous. The mutant alleles segregate with the disease in an autosomal recessive manner of inheritance [9,18]. Mutation c.1159G>A, a nucleotide change at cDNA position 1159 of the CYP1B1 gene sequence is the most prevalent homozygous mutation found in two patients (Table 2) [15,16] (Acharya et al. 2006).
Single nucleotide polymorphisms (SNP) analyses
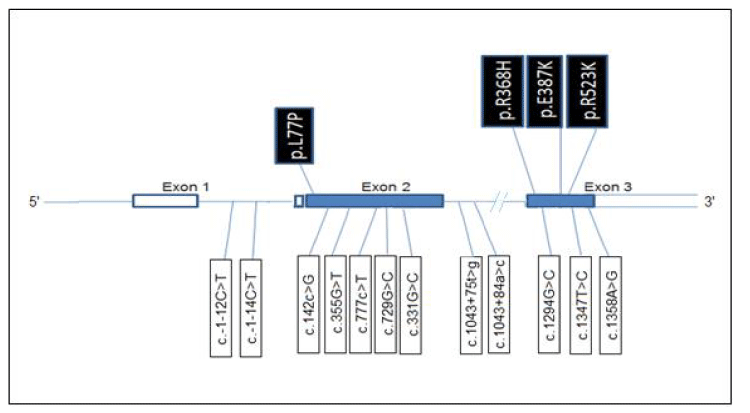
Sequencing of 19 unrelated probands with PCG detected 12 SNPs which included four intronic variants (two in intron 1 and two in intron 2), five variants in exon 2, and 3 variants in exon 3 (Table 2) [15,16] (Acharya et al. 2006). All SNPS identified in intron1 and protein coding exons 2 and 3 have been previously reported in dbSNP, while the changes found in intron 2 are novel to the best of our knowledge (Table 2) [15,16] (Acharya et al. 2006). Intronic variant c.1043+75 t>g, was the most frequent intronic change occurring in 18/19 patients, followed by intronic change c.-1-12C>T found in 8/19 patients, and c.-1-14C>T and c.1043+84a>c, each in 1/19 patient.
The most frequent exon 3variants were c.1294G>C and c.1347T>C which were each found in 16/19 patients, followed by c.1358A>G which was present in 6/19 patients.
In exon 2, c.142C>G was the most frequent SNP and was found in 9/16 patients. This was followed by c.355G>T (7/19) and c.777C>T, c.729G>C and c.331G>C (all in 1/19). From the result, it could be deduced that patients who had pathogenic mutations were homozygous for c.-1-12C>T variant. However, only three of the mutant patients had this intronic change. Mutant patients, who had coding (exonic) SNPs, had them in homozygous states except one of the compound heterozygous mutant probands (Table 4).
These four coding SNPs; R48G, A119S, V432L, N453S combined as RAVN haplotype, are inherited together as major background haplotype for identified mutations [9].
Exons are nucleic acid sequences or the expressed region of the gene, that codes for a protein. While introns are the intragenic regions or intervening sequences that do not code for proteins but are removed through (Ribonucleic Acid) RNA splicing [19] (Figure 2).
Variants found in five PCG patients
Patient JH5: c.1159G>A; p.E387K
Brief clinical details and family history: Patient JH5 is 23 years old. Mother may have had congenital glaucoma and father has cleft palate (CLP). JH5 presented with symptoms of isolated and bilateral glaucoma (PCG). JH5 also has CLP.
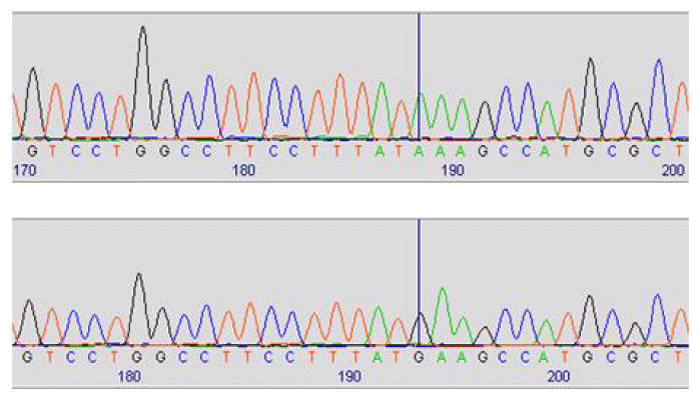
Sequencing analysis shows a previously reported homozygous missense change c.1159G>A; p.E387K in exon 3 of CYP1B1 gene. Five previously reported polymorphisms were identified in this patient (c.-1-12C>T, c.142C>G p.R48G, c.355G>T p.A119S, c.1294G>C p.V432L, c.1347T>C p.D449D) and one novel variant (c.1043+75t>g) (Figure 3).
p.E387K is a lysine to glutamine substitution that results from a homozygous 1159G>A transition. It occurs in a highly conserved region of CYP1B1 exon 3 and affects the conserved K helix region of the CYP1B1 molecule. Also, this mutation is a founder PCG mutation in the Gypsy subpopulation of Slovakia. Cleft palate results from mutations in Transcription factor AP2 (TFAP2A) gene. TFAP2A may contribute to anterior segment development.
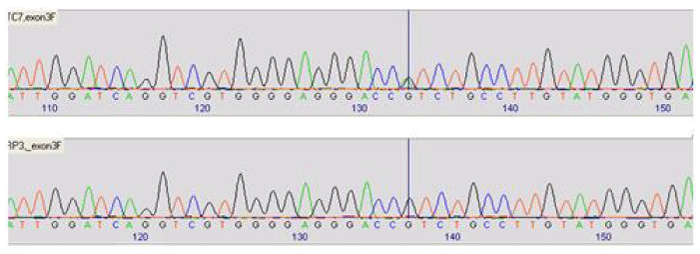
Patient TC7: c.230T>C p.L77P; c.1103G>A p.R368H
Brief clinical details and family history: TC7 is 3 years old, born prematurely and presented with bilateral congenital glaucoma.
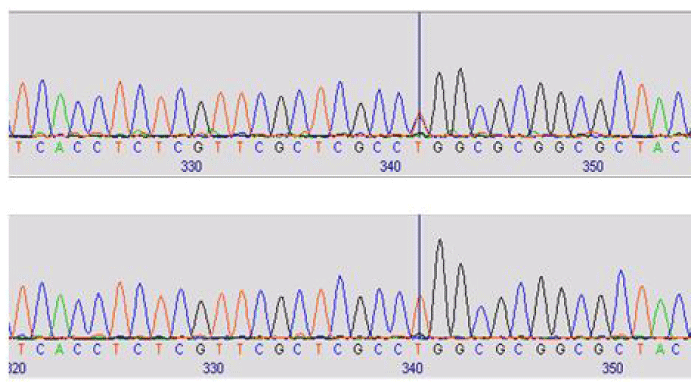
Two previously reported heterozygous missense changes c.230T>C and c.1103G>A were identified in exons 2 and 3 respectively. Three coding polymorphisms (p.N453S, p.V432L and p.D449D) and one noncoding variant (c.1043+75t>g) were identified (Figures 4,5).
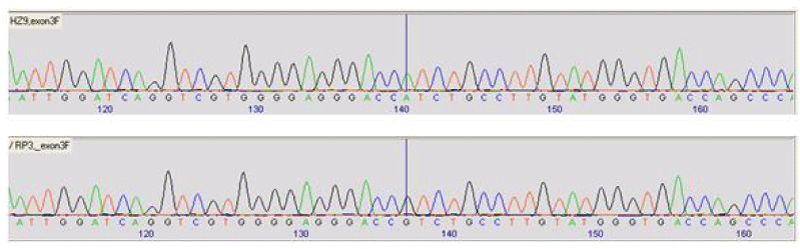
Patient HZ9: c.1103G>A; p.R368H
Brief clinical details and family history: Patient HZ9 is a 5 years old child of a consanguineous union who has congenital glaucoma, dysplastic kidneys and seizures that may be syndromic. Also, this patient has done affy array but no result was given.
Sequencing analysis shows previously reported homozygous missense change c.1103G>A; p.R368H in CYP1B1 exon 3. One novel noncoding variant (c.1043+75t>g) was also identified (Figure 6).
G>A transition at nucleotide 1103 produces an arg368-to-his (R368H) mutation found in Saudi Arabian PCG patients. This change showed incomplete penetrance and was reported to be absent in 100 Saudi Arabian control chromosomes. Also, this mutation has been reported to encode a protein with reduced, not abolished enzymatic activity.
Patient MF10: c.1159G>A p.E387K
Brief clinical details and family history: Patient MF10 is 3 years old and presented with bilateral anterior segment dysgenesis syndrome (Peter’s anomaly) manifesting as corneal clouding. This condition was present in sister and cousin, and the child is of nonconsanguineous parents.
Sequencing analysis shows previously reported homozygous missense change c.1159G>A p.E387K in CYP1B1 exon 3. Also, results show three coding SNPs and 2 noncoding variants (Table 4).
P.E387K mutation is a founder PCG mutation in the Gypsy population as previously described in patient JH5.
Patient BY17: c.1568G>A p.R523K
Brief clinical details and family history: Patient BY17 is 9 years old that presented with congenital glaucoma, heart murmur delay and Rieger phenotype. This patient has terminal deletion of a gene on Chromosome 6 (result not shown).
Sequencing analysis showed a single heterozygous CYP1B1 mutation on exon 3. In addition, two coding and one noncoding variant were found in this patient (Figure 7).
Congenital glaucoma in this patient was not due to CYP1B1 gene mutation. As stated earlier, glaucoma-associated Axen-field Rieger syndrome is due to deletion of gene located in chromosome 6p25 (forkhead Box C1 gene). This is responsible for the glaucoma in this patient.
Discussion
CYP1B1
gene mutations are the major molecular cause of PCG [20]. PCG is a genetically heterogeneous phenotype [13]. In this research, the genetic analysis of the coding exons 2 & 3 in CYP1B1 gene (Ensembl transcript ESNT00000260630) of 15 unrelated PCG cases is carried out. It shows that about 4 (26.67%) out of 15 probands had at least one PCG-causing mutation previously identified in scientific research. In the four patients having mutations, three had homozygous mutations while one showed compound heterozygosity.In addition, one patient (BY17) had a single heterozygous mutation in CYP1B1 and a terminal deletion in forkhead transcription factor gene (FKHL 7). The deletion in the short arm of chromosome 6 (6p25) is responsible for Axenfeld-Rieger Syndrome (ARS) and glaucoma [21]. Patient (TC7) was born prematurely. And it is known that anterior segment anomalies can be present in premature babies [22,23].
The pick-up rate (26.67%) may be because sequencing was limited to CYP1B1 coding exons leaving the promoter or non-coding regions [8]. Other reasons could be that the PCG phenotype may be caused by mutation in other PCG-associated loci (such as GLC3B, GLC3C or GLC3D (LTBP2) [8]. Pathogenic mutation in the LTBP2 gene associated with PCG has been reported in Pakistani, European Gypsy and Iranian probands [24,25]. Moreover, it has been suggested that a pathogenic mutation in MYOC gene may cause PCG with a CYP1B1 mutation via a digenic mode [26]. However, this finding is not established. Mutation in other unknown genes may be responsible for PCG in the CYP1B1 negative patients [8].
The rest fourteen patients were CYP1B1 negative. BY17 has ARS, so can be discounted. Although exons 2 in four probands (JH5, MF10, CS15 and JL21) was not fully sequenced, homozygous pathogenic mutations were found in two (JH5 and MF 10) (Table 2) [15,16] (Acharya et al. 2006).
From our results, PCG showed allelic heterogeneity in the patients (JH5 and MF10) associated with homozygosity for 2 different CYP1B1 mutations and on patient (TC7) showing compound heterozygosity in two distinct CYP1B1 mutations. Allelic heterogeneity explains the molecular contribution for a uniform clinical manifestation of probands, and homozygous and compound heterozygous explains the autosomal recessive nature of PCG [15,27].
p.R368H (found in TC7 and HZ9) is a hypomorphic mutation located in the conserved core structure (CCS), J helix region of the CYP1B1 protein [16]. This causes a decline in CYP1B1 enzymatic activity (~ 20% of wild type protein) or decrease in protein stability [27]. In a study by Li et al. [20], p.R368H was mostly found in white Europeans [20].
Mutation p.E387K (found in JH5 and MF10) is most common among Slovakian Gypsies, less in caucasians (4.90%), present in Amish population and absent in Asians and middle easterners [18,20]. For effective genetic screening in PCG patients, it is necessary to find out the common and founder mutations in a given population [21].
E387K is the founder mutation that accounts for 79.63% of CYP1B1 gene mutations [15]. The high rate of consanguinity (especially cousin to cousin marriages) and high coefficient of inbreeding in the Middle Eastern and Gypsy populations explains the high occurrence of the E387K mutation [20].
The amino acid position (Glu387) in which p.E387K mutation changes glutamic acid to lysine, is a highly conserved position in all documented species and P450 enzymes [28]. Also it’s a core element located in helix K, a region suspected to be essential for proper folding and active haem binding of the CYP1B1 enzyme [19,29].
The mutation found in patient BY17 (c.1568G>A) was previously identified in Israeli Bedouin kindred and it was stated that it obliterates the DdeI restriction site and disrupts the CYP enzyme active site found in the C-terminal region of the CYP1B1 enzyme [27]. Bar-yoseph et al. [27], reports this variant that changes arginine to lysine in amino acid position 523 (p.R523K), was not found in 100 healthy individuals [27].
In our study, fourteen patients had no disease-associated mutations in the fully sequenced regions of the CYP1B1 gene. This supports the report that some unidentified molecular etiology is behind the PCG in some patients [27].
Residue change from leucine (L) to Proline (P) at position 77 (p.L77P), results from c.230T>C missense mutation. This previously reported mutation (found in patient TC7) occurs in conserved position. It has been reported to be associated with PCG in Saudi families [16].
Information from this study shows that early genetic testing is pertinent to determine the carrier status of individuals and their phenotype [20]. Late presentation of PCG is associated with profound visual impairment in children [21].
Having identified CYP1B1 negative patients, future studies can be undertaken to determine other genetic causes of PCG by whole exome sequencing of these subjects.
Furthermore, the absence of CYP1B1 mutations in some PCG patients supports that another unidentified gene is mutated. The residual level of CYP1B1 activity is modulated by the presence of modifier genes [9].
Conclusions
The number of CYP1B1 negative (11/15) probands was higher than the CYP1B1 positive (4/15) patients. Some mutations may have been missed due to the genetic screening strategy applied. This is the first drawback of this study. Screening was not extended up to the regulatory sequences in the upstream or downstream regions in the introns although bit of introns were sequenced due to primer design [9].
The noncoding regions (including exon 1) may have the disease-related variants whereas this study screened only protein coding exons 2 and 3 of the gene [8]. For accurate genetic diagnosis of PCG, all three exons need to be sequenced [9].
Secondly, the small sample size (19) prevents conclusions to be confidently made; confirmation of result is therefore needed with larger number of subjects [20].
In conclusion, the analysis of CYP1B1 genotypes of 19 unrelated patients is reported in this study. A relatively low proportion of our subjects (~ 27%) tested positive for PCG-related CYP1B1 mutations demonstrate the need to identify other PCG-causing genes.
Exome sequencing of CYP1B1 negative patients to detect new genes mutated in PCG.
Co-segregation analysis of other unaffected and affected family members may be carried in the future.
- Mandal AK, Chakrabarti D (2011) Update on congenital glaucoma. Indian J Ophthalmol 59:148-57. Link: https://tinyurl.com/y8fe3fws
- Shaffer RN, Weiss DI (1970) Infantile glaucoma: diagnosis and differential diagnosis. Congenital and Pediatric Glaucomas. CV Mosby St. Louis.
- Ritch R, Shields MB, Krupin T (1996) The glaucomas: glaucoma therapy, 2nd ed. Mosby-year Book Inc.
- Anderson DR (1981) The development of the trabecular meshwork and its abnormality in primary infantile glaucoma. Trans Am Ophthalmol Soc 79: 458-485. Link: https://tinyurl.com/y9qyb8ee
- deLuise VP, Anderson DR (1983) Primary infantile glaucoma (congenital glaucoma). Surv Ophthalmol 28: 1-19. Link: https://tinyurl.com/yadvtdg3
- Bejjani BA, Xu L, Armstrong D, Lupski JR, Reneker LW (2002) Expression Patterns of Cytochrome P4501B1 in FVB/N Mouse Eyes. EExp Eye Res 75: 249-257. Link: https://tinyurl.com/ydxeepop
- Pagon RA, Bird TD, Dolan CR, Stephens K, Adam MP, et al. (2004) Primary Congenital Glaucoma.
- Kim HJ, Suh W, Park SC, Kim CY, Park KH, et al. (2011) Mutation spectrum of CYP1B1 and MYOC genes in Korean patients with primary congenital glaucoma. Molecular vision 17: 2093-2101. Link: https://tinyurl.com/yaksxz7x
- López-Garrido MP, Medina-Trillo C, Morales L, Garcia-Feijoo J, Martínez-de-la-Casa JM, et al. (2012) Genotypes in Primary Congenital and Non dominant Juvenile Glaucoma. Ophthalmology.
- Genĉík A (1989) Epidemiology and genetics of primary congenital glaucoma in Slovakia. Description of a form of primary congenital glaucoma in gypsies with autosomal-recessive inheritance and complete penetrance. Developments in ophthalmology 16: 76-155. Link: https://tinyurl.com/ybjzm724
- Gencík A, Gencíková A, Gerinec A (1980) Genetic heterogeneity of Congenital glaucoma. Clin Genet 17: 241-248. Link: https://tinyurl.com/y9aqbh82
- Kaur K, Reddy ABM, Mukhopadhyay A, Mandal AK, Hasnain SE, et al.(2005) Myocilin gene implicated in primary congenital glaucoma. Clinical Genetics 67: 335-340. Link: https://tinyurl.com/ybe8aqwe
- Stoilov I, Akarsu AN, Sarfarazi M (1997) Identification of three different truncating mutations in cytochrome P4501B1 (CYP1B1) as the principal cause of primary congenital glaucoma (Buphthalmos) in families linked to the GLC3A locus on chromosome 2p21. Hum Mol Genet 6: 641-647. Link: https://tinyurl.com/yc6zt6a2
- Calugaru D (2010) Mendelian molecular genetics in glaucoma. Oftalmologia 54: 8-20. Link: https://tinyurl.com/ya8vju5c
- Plášilová M, Stoilov I, Sarfarazi M, Kádasi L, Feráková E, et al. (1999) Identification of a single ancestral CYP1B1 mutation in Slovak Gypsies (Roms) affected with primary congenital glaucoma. J Med Genet 36: 290-294. Link: https://tinyurl.com/y9sdn5lg
- Bejjani BA, Stockton DW, Lewis RA, Tomey KF, Dueker DK, et al. (2000) Multiple CYP1B1 mutations and incomplete penetrance in an inbred population segregating primary congenital glaucoma suggest frequent de novo events and a dominant modifier locus. Human Molecular Genetics 9: 367-374. Link: https://tinyurl.com/y8jqualy
- Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors.Proceedings of the National Academy of Sciences 74: 5463-5467. Link: https://tinyurl.com/ydxrsxd2
- Martin SN, Sutherland J, Levin AV, Klose R, Priston M, et al. (2000) Molecular characterisation of congenital glaucoma in a consanguineous Canadian community: a step towards preventing glaucoma related blindness. J Med Genet 37: 422-427. Link: https://tinyurl.com/yb8pwck7
- Berk AJ (2006) Discovery of RNA splicing and genes in pieces. Proc Natl Acad Sci USA 113: 801-805. Link: https://tinyurl.com/ycrcycyz
- Li N, Zhou Y, Du L, Wei M, Chen X (2011) Overview of Cytochrome P450 1B1 gene mutations in patients with primary congenital glaucoma. Experimental Eye Research 93: 572-579. Link: https://tinyurl.com/ya8dnvuw
- Nishimura DY, Swiderski RE, Alward WL, Searby CC, Patil SR, et al. (1998) the forkhead transcription factor gene FKHL7 is responsible for glaucoma phenotypes which maps to 6p25. Nat Genet 19: 140-147. Link: https://tinyurl.com/yd24tkjs
- Worst JGF (1966) The Pathogenesis of Congenital Glaucoma. Royal Vangorcum, Assen. Netherlands.
- Kwitko ML (1973) Glaucoma in Infants and Children. Meredith Corporation, New York.
- Ali M, McKibbin M, Booth A, Parry DA, Jain P, et al. (2009) Null Mutations in LTBP2 Cause Primary Congenital Glaucoma. American Journal of Human Genetics 84: 664-671. Link: https://tinyurl.com/y7dm5rzn
- Narooie-Nejad M, Paylakhi SH, Shojaee S, Fazlali Z, Rezaei Kanavi M, et al. (2009) Loss of function mutations in the gene encoding latent transforming growth factor beta binding protein 2, LTBP2, cause primary congenital glaucoma. Human molecular genetics 18: 3969-3977. Link: https://tinyurl.com/yav4kn9z
- Vincent AL, Billingsley G, Buys Y, Levin AV, Priston M, et al. (2002) Digenic Inheritance of Early-Onset Glaucoma: CYP1B1, a Potential Modifier Gene. Am J Hum Genet 70: 448-460. Link: https://tinyurl.com/yap795yq
- Bar-Yosef U, Levy J, Elbedour K, Ofir R, Carmi R, et al. (2010) Congenital Glaucoma CYP1B1 Mutations in Israeli Bedouin Kindreds. J Glaucoma 19: 35-38. Link: https://tinyurl.com/ybtuynto
- López-Garrido MP, Blanco-Marchite C, Sánchez-Sánchez F, López-Sánchez E, Chaqués-Alepuz V, et al. (2010) Functional analysis of CYP1B1 mutations and association of heterozygous hypomorphic alleles with primary open‐angle glaucoma. Clin Genet 77: 70-78. Link: https://tinyurl.com/yc6vkhq3
- Stoilov I, Akarsu AN, Alozie I, Child A, Barsoum-Homsy M, et al. (1998) Sequence analysis and homology modeling suggest that primary congenital glaucoma on 2p21 results from mutations disrupting either the hinge region or the conserved core structures of cytochrome P4501B1. Am J Hum Genet 62: 573-584. Link: https://tinyurl.com/yco8b4z6
- Kim HS, Hong SJ, LeDoux MS, Kim KS (2001) Regulation of the tyrosine hydroxylase and dopamine β‐hydroxylase genes by the transcription factor AP‐2. J Neurochem 76: 280-294. Link: https://tinyurl.com/ycmqhmsk
- Ben-Zion I, Bogale A, Moore DB, Helveston EM (2010) Bilateral primary congenital glaucoma in monozygotic twins. J Pediatr Ophthalmol Strabismus 47: 124-126. Link: https://tinyurl.com/yajyv2qq
- Nebert D W (1990) DRUG-METABOLISM - GROWTH SIGNAL PATHWAYS. Nature 347: 709-710. Link: https://tinyurl.com/y7upjhqn
- Nebert DW, Russell DW (2002) Clinical importance of the cytochromes P450. The Lancet 360: 1155-1162. Link: https://tinyurl.com/y7hyx4h5
- Schwartzman ML, Balazy M, Masferrer J, Abraham NG, McGiff JC, et al. (1987) 12 (R)-hydroxyicosatetraenoic acid: a cytochrome-P450-dependent arachidonate metabolite that inhibits Na+, K+-ATPase in the cornea. Proceedings of the National Academy of Sciences 84: 8125-8129. Link: https://tinyurl.com/yb8m2qug
- Kupfer C, Kaiser-Kupfer MI (1978) New hypothesis of developmental anomalies of the anterior chamber associated with glaucoma. Transactions of the ophthalmological societies of the United Kingdom 98:213. Link: https://tinyurl.com/y75zq5nv
- Vincent A, Billingsley G, Priston M, Williams-Lyn D, Sutherland J, et al. (2001) Phenotypic heterogeneity of CYP1B1: mutations in a patient with Peters' anomaly. J Med Genet 38: 324-326. Link: https://tinyurl.com/ybu8vnmm
- Dimasi DP, Hewitt AW, Straga T, Pater J, MacKinnon JR, et al. (2007) Prevalence of CYP1B1 mutations in Australian patients with primary congenital glaucoma. Clin Genet 72: 255-260. Link: https://tinyurl.com/ybxthjbs
- Stoilov IR, Costa VP, Vasconcellos JP, Melo MB, Betinjane AJ, et al. (2002) Molecular genetics of primary congenital glaucoma in Brazil. Invest Ophthalmol Vis Sci 43: 1820-1827. Link: https://tinyurl.com/ycdgdrke
- Chavarria-Soley G, Sticht H, Aklillu E, Ingelman-Sundberg M, Pasutto F, et al. (2008) Mutations in CYP1B1 cause primary congenital glaucoma by reduction of either activity or abundance of the enzyme. Human mutation 29: 1147-1153. Link: https://tinyurl.com/yb94l5je
- Akarsu AN, Turacli ME, Aktan SG, Barsoum-Homsy M, Chevrette L, et al. (1996) A second locus (GLC3B) for primary congenital glaucoma (Buphthalmos) maps to the 1p36 region. Human Molecular Genetics 5:1199-1203. Link: https://tinyurl.com/yc83c2nn
- Stoilov IR, Sarfarazi M (2002) The Third Genetic Locus (GLC3C) for Primary Congenital Glaucoma (PCG) Maps to Chromosome 14q24.3. ARVO Annual Meeting Abstract Search and Program Planner 43: 3015. Link: https://tinyurl.com/y8gjn9nh
- Sutter TR, Tang YM, Hayes CL, Wo YY, Jabs EW, et al. (1994) Complete cDNA sequence of a human dioxin-inducible mRNA identifies a new gene subfamily of cytochrome P450 that maps to chromosome 2. J Biol Chem 269: 13092-13099. Link: https://tinyurl.com/yd2d2tny
- Tang YM, Wo YY, Stewart J, Hawkins AL, Griffin CA, et al. (1996) Isolation and characterization of the human cytochrome P450 CYP1B1 gene. J Biol Chem 271: 28324-28330. Link: https://tinyurl.com/y9cpcxwp
- Graham-Lorence SE, Peterson JA (1996) Structural alignments of P450s and extrapolations to the unknown. Methods in enzymology 272: 315-326. Link: https://tinyurl.com/yaftokf2

Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
