Open Journal of Bioinformatics and Biostatistics
Bioinformatic analysis of metal transportomes from Mycobacteria Sp.
Shanti Kumari Lunavat2, Jai Satya Gowri Gogada2, Surya Satyanarayana Singh2 and Raghu Gogada1,2*
2Department of Biochemistry, University Collage of Science, Osmania University, Hyderabad-500007, Telangana, India
Cite this as
Shanti Kumari L, Jai Satya Gowri G, Surya Satyanarayana S, Raghu G (2021) Bioinformatic analysis of metal transportomes from Mycobacteria Sp. Open J Bioinform Biostat 5(1): 014-022. DOI: 10.17352/ojbb.000011Copyright
© 2021 Shanti Kumari L, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Mycobacterium is immovable induce aerobic, acid-fast gram-positive bacilli with high genomic content (59-66%). In the operon structure frequently establish for the genes of three molecular components: the ABC-binding protein, the membrane protein, and the substrate-binding protein, the rates of multidrug resistant and metal ions. The main objective of this study was to analyze the metal ions from five Mycobacterium species and to identify the metal transporters with “Genomic Island” associated features, in-silico analysis allowed identification of metal and drug transporters, phylogenetic analysis, genomic island path analysis, prediction of interacting metal ions, 3D structure, domain analysis and for the NiCoT metal transporter from Mycobacterium tuberculosis. These data are the first results of a big frame project that aims to accelerate the prioritizing of gene candidates that control element accumulation by taking advantage of high-throughput. The present in-silico study reveals the complete suite of NiCoT Metal Transporter in Mycobacterium tuberculosis H37Rv, which is involved in urease enzyme activity and biological function. The STRING analysis defines that the functional partners involved in transport of metal ions. While high expression yields of membrane proteins remain significant bottleneck for many proteins.
Introduction
In recent years the complete genome sequences of versatile organisms from the three domains of life are rapidly accumulating. It is now possible to attempt to reconstruct and analyze a complete set of biochemical reaction pathways that an organism adopts, especially on the transport, synthesis, and degradation of specific chemical compounds [1]. Through comparative studies, metal ions play a life preserve role in prokaryotic metabolism [1-3]. Biological management of metal ions is skilled by a complex interplay between metal ion and drug transporters (transmembrane importers, transmembrane exporters) and their regulatory components [1-5]. Mycobacteria belong to the family Mycobacteriaceae and are members of the CMN group (Corynebacteria, Mycobacteria and Nocardia). The families Mycobacteriaceae are gram-positive, immobile, catalase-positive have a rod like to filamentous morphology and it could be pleomorphic. As a group, they produce characteristic long chain fatty acids termed mycolic acids. Mycobacteria are acid-fast rods of variable appearance, approximately 0.2 - 0.6 by 1-10 mm. The genus Mycobacterium consists of 127 species according up to the minute approved list of bacterial species [3]. Mycobacteria arranged into four groups according to the Runyon classification: a) Photochromogens: slow growers and form pigment when exposed to light. (e.g.: M. kansasii, M. marinum, M. simiae). b) Scotochromogens: slow growers and form pigment in the dark (e.g.: M. scrofulaceum, M. szulgai, M. gordonae). c) Non-photochromogens: slow growers and not pigmented (e.g.: M. malmoense, M. xenopi, M. avium-complex, M. ulcerans, M. haemophilum). d) Rapid growers: fast growers (e.g.: M. fortuitum, M. chelonae, M. abscessus). Most slow-growing species have been associated with disease in humans while only few species of group 4 are disease associated [6,7]. The identification of a new species was conventionally based on the description of the Runyon classification, the biochemical properties of the strain(s) and the degree of DNA-DNA hybridization. Taxonomically, Mycobacteria are the single genus within the family of Mycobacteriaceae, in the order Actinomycetales. It includes pre-nominal micro-organisms and they are traditionally differentiated on the basis of phenotypic characteristics, culture properties that help to separate among various species of Mycobacteria. It is also a leading cause of infection in various domesticated animals and wildlife. The Mycobacterial cell envelope, which is an analyzable tripartite structure containing a high proportion of lipids (approximately 30% to 40% of the total weight) could play an essential role in the adaptation of Mycobacteria to intracellular growth and survival, immune modulation and drug resistance [7]. The availability of complete genome sequences of five species namely M. avium K10, M. leprae AF2122, M. bovis 97, M. tuberculosis H37Rv, M. smegmatis str. MC2155 provided an opportunity to analyze the metal transporters and multidrug transporters [8-22]. Secondary transporters from the NiCoT family are able to uptake either both Ni and Co, or prefer only Ni ions. NiCoT’s are widespread among bacteria and found in some Archaea and fungi. Substrate preferences correlate with the genomic localization of NiCoT genes adjacent to clusters of Ni/Co - dependent enzymes and enzymes of B12 biosynthesis, as well as with the presence of Ni or B12 regulatory sites upstream [4,5,23-35].
Methods
Compilation of metal and multidrug transporters
The genome sequence information of five sequenced species M. avium K-10, M. leprae AF2122, M. bovis 97, M. tuberculosis H37Rv, M. smegmatis str. MC2155 in this study were selected from transport database (33). Transport database is a relational database describing details of a comprehensive IUBMB approved classification system for transport proteins known as classification of enzymes. TCDB is freely web accessible at http://www.tcdb.org, curated relational database and also containing protein sequence, classification, structural, functional and evolutionary information about transport systems from a variety of living organisms. It offers several tools specifically designed for analyzing the unique characteristics of transport proteins and serves as a genome transporter annotation tool. In order to search for homologs of a transporter family the best-composed hit was used in a subsequent BLASTP search against the five species whole protein sequences and retrieved the members. From the compiled sequences phylogenetic trees were constructed for substrate specific transporters individually and through analyzing the aligned sequences conserved domains for individual metal ion and drug transporter were reported, protein search was also carried out with retrieved homolog’s using CLC sequencer database version 6.9.1 [10]. For some of the specific nickel secondary transporter and cobalt transporters orthologous and paralogous sequences were obtained using KEGG database and phylogenetic trees were generated for them (KEGG database). Transmembrane helix prediction for the membrane transporters was performed using Transmembrane Hidden Markov Model (TMHMM) version 2.0. Genomic location and gene organization search was carried out using ISLAND PATH analysis [4,5,12,35].
Protein sequence analysis
CLC workbench has employed to analyze the metal transporter and multidrug transporter proteins [10]. The protein sequences were collected and aligned to identify regions of similarity that may be a consequence of functional, structural or evolutionary relationships between the sequences. Conserved domains are identified by using Motif Search in KEGG database. The conserved motif in the ATP dependent transporter’s membrane domain “LSGGQ” has been identified. This domain is the signature sequence for the ABC transporter proteins [4,5,10].
Phylogenetic analysis
Phylogenetic analysis is an illustration of the evolutionary relationships among a group of organisms. It was performed with the characterized prototypes using CLC protein workbench. For this analysis, Multiple Sequence Alignments (MSA) were produced using progressive alignment algorithm. The generated pair wise alignments were used for finding the evolutionary distance between the pairs. Pair wise distances thus calculated was used to create a Phylogenetic tree-employing Neighbor Joining (NJ) algorithm with 1000 bootstrap replicates [5,10].
Island path analysis
Mycobacteria species are ecologically diversified organisms habituated to grow in host-associated environmental conditions and some are multiple. In the five species named above except Mycobacterium leprae all are host-associated and it is multiple. Organisms to get acquainted to specific niche, they need to meet the requirements to survive in that conditions. Horizontal gene transfer mechanism is involved in the achievement of essential needs. Island Path Analysis was used for the detection of metal and drug transporters acquired through HGT. After generating the complete inventory of metal and drug transporters, we inspected the genomes of five Mycobacterium species with island path software (IPA version 1.0 tool) for the identification of those transporters located in GI’s or exhibiting GI associated features like anomalous %G+C, dinucleotide bias above 1 STD DEV, presence of RNA genes (tRNA, rRNA genes) and mobility genes (transposons, insertion sequences). A pre-nominal GI can be identified with certainty by the presence of eight or more consecutive ORF’s with dinucleotide bias alone or dinucleotide bias plus a mobility gene in proximity [1].
Identification of metal ion transporters and their sub cellular localization
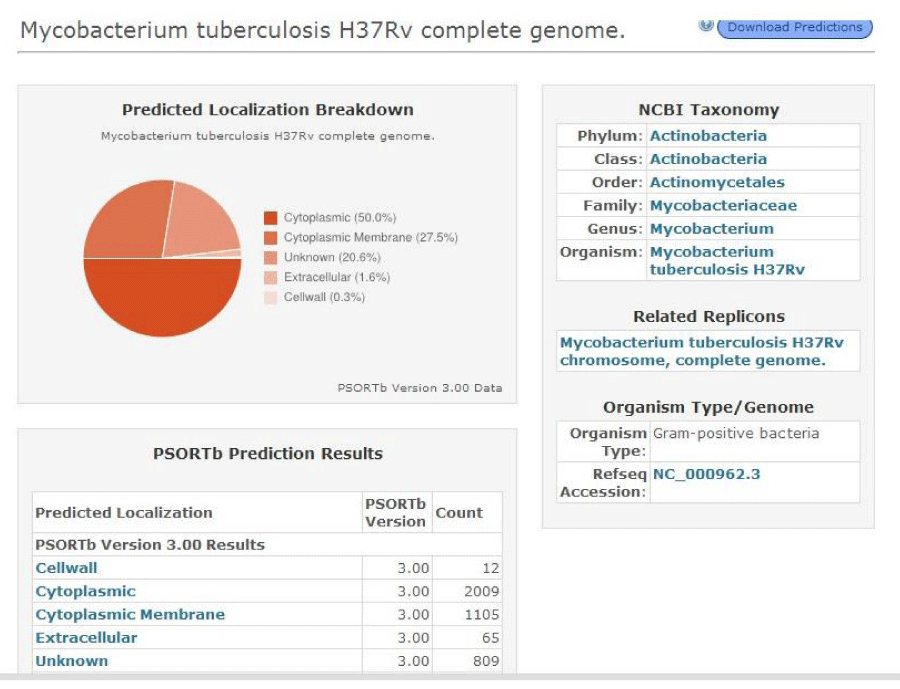
The protein transporter has transmembrane helices which are identified using a tool TMHMM (Trans Membrane prediction using Hidden Markov Models) an option present in Transport DB. The transporters with transmembrane helices are then subjected for the identification of protein sub cellular localization using PSORTdb (http://db.psort.org/). It is a web accessible database of SCL for bacteria that contains both information determined through laboratory experimentation and computational predictions [12].
Prediction of interaction and interacting partners of the metal ion transporters
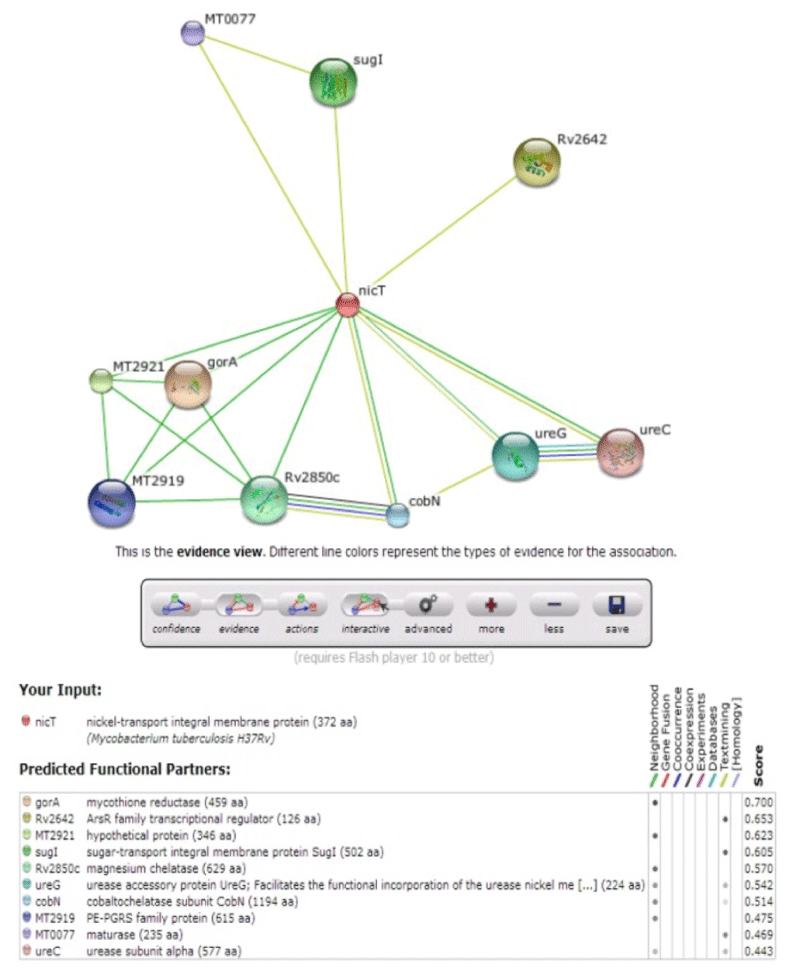
The Nickel & Cobalt metal transporters are the major constituents in urease enzyme & many biological functions. The protein-protein interactions are studied using STRING database (http://string-db.org/). It is pre-computed global resource for the exploration and analysis of the associations. Since the evidence differs conceptually, and the number of predicted interactions is very large, it is essential to be able to assess and compare the significance of individual predictions. Thus, STRING contains a unique scoring-framework based on benchmarks of the different types of associations against a common reference set, integrated in a single confidence score per prediction [5,14].
Prediction of 3D structure and domain analysis
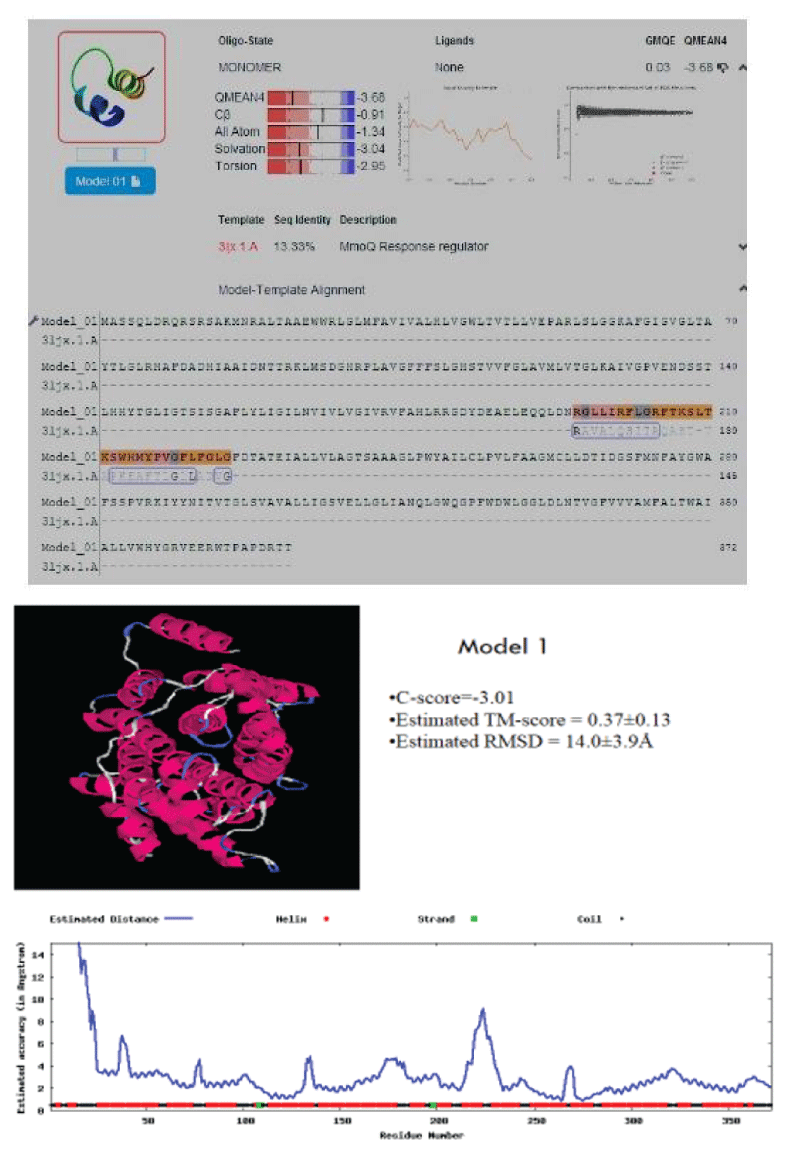
After STRING analysis, the NiCoT transporter which is involved in transport of Nickel efflux are subjected to Homology Modeling and their 3D models are generated by selecting the reliable template using Swiss-Model (http:// swissmodel.expasy.org/) [15]. The SWISS-MODEL template library provides annotation of quaternary structure and essential ligands and cofactors to allow for building of complete structural models, including their oligomeric structure. The 3D structures that were built are subjected for verification using RAMPAGE (http://mordred. bioc.cam.ac.uk) and active domains are analyzed using ProDom (http://prodom.prabi.fr) [1].
Results
From genome to metal and drug transportome
Based on the global features of five Mycobacteria genomes we could draw a comparison among the genome size, total number of genes, transporter proteins, G+C content (%), total number of metal transporters, total number of drug transporters (Table 1). As shown in the table 1, M. smegmatis has largest genome size and M. leprae has smallest genome size in comparison. Among the total metal and drug transporters M. smegmatis has the highest number of proteins when compared with M. leprae.
Based on membrane transporter database (Transport DB), we have compiled the metal and drug transportomes for the alkaline earth metal (Mg2+), transition metal ions (Zn2+, Mn2+, Cu2+, Ni2+, Co2+), and heavy metal (Cd2+) in five species of Mycobacteria (Table 2).
From the above analysis, as shown in Table 3, we were reported that M. smegmatis has the highest number of ATP dependent transporters (12 ATP dependent metal & 28 drug transporters) among the group and secondary metal transporters are high in M. avium [7] and drug transporters in M. smegmatis (128). Ion channels and also the unclassified transporters are very less in number in all the group members and they are identified in metal transporters.
Salient features of metal and multidrug transportomes
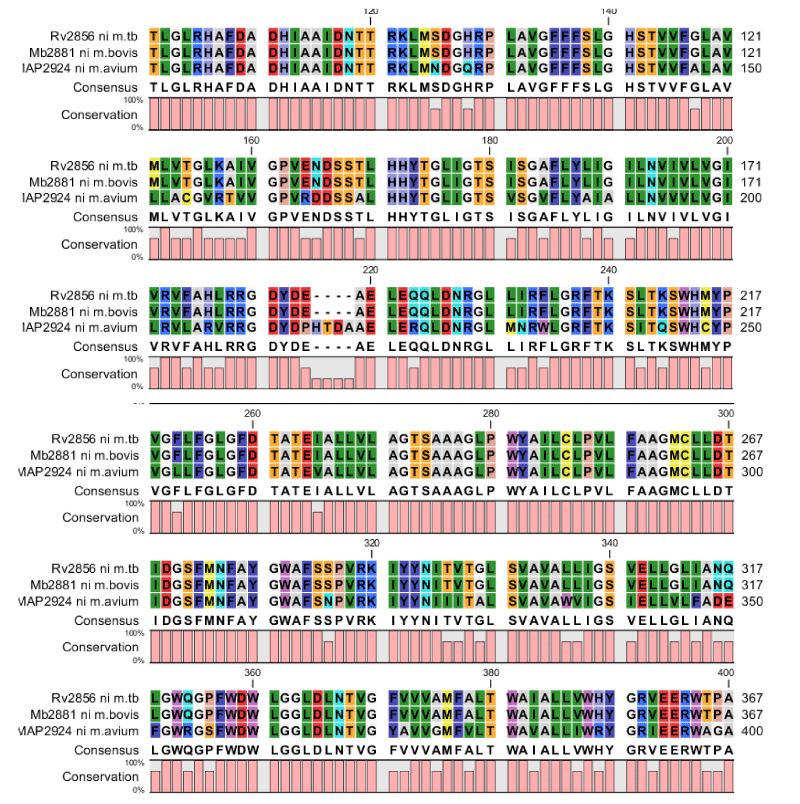
Apart from the Transporter Database the protein information is provided in Mycobacterium database as well as in Transport DB of Tuberculosis Database. The up to date information is maintained and updated if any new protein is identified. The metal ion transporters and multidrug transporters are identified in all five species mentioned above and conserved motif domains are also identified for these respective species. Dataset cataloguing and multiple sequence alignment of the sequences helped us to find the unique signature sequences (Figure 1). The major finding of our study is that there are only three Nickel transporters from three species found among all the organisms from the ground and it is secondary transporter belongs to Ni2+Co2+ transporter family. We have mainly focused on Nickel-Cobalt metal ion transport and multidrug transporters. The nickel-cobalt metal ion is identified in three species of selected five species (Figure 2). It is identified in M. bovis (Mb2881), M. tuberculosis (Rv2856) and M. avium (MAP2924). As of now up to date the main motif is identified in Nickel-Cobalt is “HAFDADH” in second transmembrane helix. But when compared to these three species we have identified the main motif “HAFDADH” in third transmembrane helix and also other three conserved motif regions are identified in sixth, seventh and eighth peaks of transmembrane helix. The proteins were collected and by using Motif search from KEGG database motifs are identified and by using TMHMM version 2 software the conserved domain motifs are confirmed by analyzing the transmembrane helix regions (Figure 1). “VGFLFGLGFD” – in sixth peak, “IDGSFMNAYGWAFS” – in seventh peak and “LGGLDLNTVG”- in eighth peak. In fourth and fifth transmembrane helix the conserved motifs are identified but around 95% identity. In these two conserved domain motifs one of the amino acid sequence is 99% identity. “SSTLHHYTG”- in fourth peak and “LEQQLDNRGL” – in fifth peak.
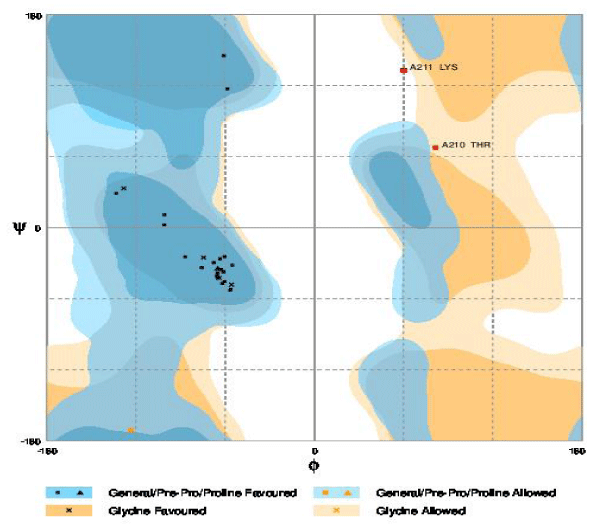
The multidrug transporters are aligned using CLC Workbench Software and ClustalW and Phylogenetic tree is also constructed (Figure 2). Horizontal gene transfer effect plays an important role in the acquisition of the requirements to adapt to specific niche conditions. Island path analysis tool is used to analyze Genomic island associated features of metal ion transporters for 5 Mycobacterial sp. Thus, island path analyzer gave us the clear idea on the number of metal ion transporters acquired through HGT mechanism in the Mycobacterial sp. [34]. The NiCoT (Rv2856) metal transporter with its sub cellular localization is identified from Transport DB and PSORTdb respectively (Figure 3). From the above-mentioned data, the protein is subjected to STRING analysis and their interactions and interacting partners are identified. The interactions indicate their functional protein partners are involved in the mechanism of transport of metal ion are interacting with each other (Figure 4). STRING database analyzed protein is subjected to Homology Modeling & 3D structures are relied which are generated using the template from Swiss Model tool (Figure 5). The structures are validated using RAMPAGE (Figure 6) and its function & domains are identified using PRODOM. From this analysis it is clear that the NiCoT protein transports the metal ion as their functional domain is Secondary Transporter.
Discussions
In this study we have performed a dataset cataloguing for 69 metal and 243 drug transporter proteins from five Mycobacterial sp. This is the comprehensive genomic comparison of metal transporters, providing potentially important insights into the fundamental molecular aspects and novel facets of Mycobacterial metal transporters. Transition metals nickel and cobalt are essential components of many metalloenzymes. Ni-dependent enzymes are urease, [NiFe] hydrogenase [Ni] superoxide dismutase, CO dehydrogenase, and methyl-CoM reductase. In the form of coenzyme B12, cobalt plays a number of crucial roles in many biological functions. Also, there are some noncorrin-cobalt-containing enzymes (e.g. nitrile hydratase). Synthesis of Ni / Co enzymes and coenzyme B12 requires high-affinity uptake of the metal ions from natural environments where they are available only in trace amounts. Ni and Co uptake in bacteria is mediated by various secondary transporters and by at least two different ATP-Binding Cassette (ABC) systems. It is thought to be involved in transport of nickel across the membrane responsible for translocation of substrate across membrane. For the urease enzyme activity nickel metal ion is used as a co-factor and vitamin B12 enzyme activity cobalt ion is used as a co-factor. The present in-silico study reveals the complete suite of NiCoT Metal Transporter in M. tuberculosis H37Rv, which is involved in urease enzyme activity and biological function. The STRING analysis defines that the functional partners involved in transport of metal ions.
The authors thank to the Department of Science and technology [INSPIRE Faculty Award (DST) IFA12-LSPA-11to GR].
- Haritha A, Rodrigue A, Mohan PM (2008) A comparative analysis of metal transportomes from metabolically versatile Pseudomonas. BMC Research Notes 1: 88. Link: https://bit.ly/3h4Qmub
- Tomii KT, Kanehisa M (1998) A Comparative Analysis of ABC Transporters in Complete Microbial Genomes. Genome Res 8: 1048-1059. Link: https://bit.ly/2VmdixF
- Barsom EK, Hatfull GF (1997) A putative ABC-transport operon of Mycobacterium. Gene 185: 127–132. Link: https://bit.ly/3tjBbSZ
- Shanti KL, Jai SGG, Praveen B, Puspashree P, Surya SS, et al. (2020) Metal and Drug Transportomes Analysis from Cyanobacterial sp., Indian Journal of Natural Sciences 10: 23945-23951.
- Shanti KL, Praveen B, Surya SS, Jai SGG, Raghu G (2020) NiCoT Transporters from Microorganisms for Metal Transporter Studies-Mini Review. Indian Journal of Natural Sciences 10: 24269-24287.
- Rastogi N, Legrand E, Sola C (2001) The Mycobacteria: an introduction to nomenclature and pathogenesis. Rev Sci Tech 20: 21-54. Link: https://bit.ly/3jONlQJ
- Wayne LG, Good RC, Tsang A, Butler R, Dawson D, et al. (1993) Serovar determination and molecular taxonomic correlation in Mycobacterium avium,Mycobacterium intracellulare, and Mycobacterium scrofulaceum: a cooperative study of the International Working Group on Mycobacterial Taxonomy. Int J Syst Bacteriol 43: 482-489. Link: https://bit.ly/3BK6AAY
- Saier MH, Tran CV, Barabote RD (2006) TCDB: The Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res 34: D181–D186. Link: https://bit.ly/3narYLx
- Webb MR (2003) Mycobacterial ABC Transport System: Structure of the Primary Phosphate Receptor. Structure 11: 736-738. Link: https://bit.ly/3nai250
- CLC Sequence Viewer User Manual. Link: www.clcbio.com.
- Schmalstieg AM, Srivastava S, Belkaya S, Deshpande D, Meek C, et al. (2012) The Antibiotic Resistance Arrow of Time: Efflux Pump Induction Is a General First Step in the Evolution of Mycobacterial Drug Resistance. Antimicrob Agents Chemother 56: 4806-4815. Link: https://bit.ly/3BKO4sg
- Kanehisa M, Gotto S (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28: 27-30. Link: https://bit.ly/3na9ACQ
- Nies DH (1992) Resistance to Cadmium, Cobalt, Zinc, and Nickel in Microbes. Plasmid 27: 17-28. Link: https://bit.ly/3BJRPhF
- Eitinger T, Mandrand-Berthelot MA (2000), Nickel transport systems in microorganisms. Arch Microbiol 173: 1-9. Link: https://bit.ly/3haMGY9
- Hediger MA, Romero MF, Peng JB, Rolfs A, Takanaga H, et al. (2004) The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteins. Pflugers Arch 447: 465-468. Link: https://bit.ly/3E30gGX
- Kehres DG, Maguire ME (2002) Structure, properties and regulation of magnesium transport proteins. BioMetals 15: 261-270. Link: https://bit.ly/3BEHfse
- Nies DH, Silver S (1995) Ion efflux systems involved in bacterial metal resistances. Journal of Industrial Microbiology 14: 186-199. Link: https://bit.ly/2Vjeba2
- Nies DH (1999) Microbial heavy-metal resistance, Appl Microbiol Biotechnol 51: 730-750. Link: https://bit.ly/3yJRbyM
- Eitinger T, Suhr J, Moore L, Smith AC (2005) Secondary Transporters for Nickel and Cobalt Ions: Theme and Variations. BioMetals 18: 399-405. Link: https://bit.ly/2X0p9SO
- Mulrooney SB, Hausinger RP (2003) Nickel uptake and utilization by microorganisms. FEMS Microbiol Rev 27: 239-261. Link: https://bit.ly/3h6tudO
- Rodionov DA, Hebbeln P, Maurel J, Gelfand MS, Eitinger T (2020) Comparative genomic analysis of Nickel and Cobalt uptake transporters in bacteria. Characterization of a novel ABC-type transport system.
- Chakraborti PK, Bhatt K, Banerjee SK, Misra P (1999) Role of an ABC importer in Mycobacterial drug resistance. Biosci Rep 19: 293-300. Link: https://bit.ly/3tkGAcl
- Garnier T, Eiglmeier K, Camus JC, Medina N, Mansoor H, et.al. (2003) The complete genome sequence of Mycobacterium bovis. Proc Natl Acad Sci U S A 100: 7877–7882. Link: https://bit.ly/38Sb245
- Schmalstieg AM, SSrivastava, Belkaya S, Deshpande D, Meek C, Leff R, et al. (2012) The Antibiotic Resistance Arrow of Time: Efflux Pump Induction Is a General First Step in the Evolution of Mycobacterial Drug Resistance. Antimicrob Agents Chemother 56: 4806-4815. Link: https://bit.ly/38NQc6e
- Kihara D, Shimizu T, Kanehisa M (1998) Prediction of membrane proteins based on classification of transmembrane segments. Protein Eng 11: 961-970. Link: https://bit.ly/3DQ7pKq
- Kiranmayi P, Rautray S, Rachel VK, Zaveri K (2013) Identification and evaluating the role of metal ion transporters in Dienococcusradiodurans using insilico tools. 2. Link: https://bit.ly/3BK6wkI
- Mulrooney SB, Hausinger RP (2003) Nickel uptake and utilization by microorganisms. FEMS Microbiol Rev 27: 239-261. Link: https://bit.ly/3kYsMRg
- Nelson N (1999) Metal ion transporters and homeostasis. EMBO J 18: 4361-4371. Link: https://bit.ly/3BRAHqn
- Hedgier MA (1997) Membrane pemiability. The Diversity of transmembrane transport processes. Curr Opin Cell Biol 9: 543-546. Link: https://bit.ly/3zPX5zv
- Eide DJ (1998) The Molecular Biology Of Metal Ion Transport In Saccharomyces Cerevisiae. Annu Rev Nutr 18: 441-469. Link: https://bit.ly/2WV5RxU
- Radisky DC, Kaplan J (1999) Regulation of transition metal transport across the yeast plasma membrane. J Biol Chem 274: 4481-4484. Link: https://bit.ly/3h4Q8TR
- Rey S, Acab M, Gardy JL, Laird MR, deFays K, et al. (2005) PSORTdb: a protein subcellular localization database for bacteria. Nucleic Acids Res 33: D164-168. Link: https://bit.ly/3jOAw8O
- von Mering C, Huynen M, Jaeggi D, Schmidt S , Bork P, et al. (2003) STRING: a database of predicted functional associations between proteins. Nucleic Acids Res 31: 258-261. Link: https://bit.ly/3l6h7zY
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, et al. (2014) SWISS-MODEL Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42: W252-258. Link: Link: https://bit.ly/3jRbhTE
- Elbourne LDH, Tetu S, Hassan K, Paulsen IT (2017) TransportDB 2.0: a database for exploring membrane transporters in sequenced genomes from all domains of life. Nucleic Acids Res 45: D320-D324. Link: https://bit.ly/3BLLuCj
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
