Open Journal of Plant Science
Bacterial blight of Brachiaria caused by Burkholderia glumae in Colombia
Elizabeth Alvarez1* and Michael Latorre2
2Colombian Corporation for Agricultural Research (AGROSAVIA), Research center: La Selva. Km. 7, Vía Rionegro - Las Palmas, Colombia
Cite this as
Alvarez E, Latorre M (2023) Bacterial blight of Brachiaria caused by Burkholderia glumae in Colombia. Open J Plant Sci 8(1): 010-019. DOI: 10.17352/ojps.000052Copyright
© 2023 Alvarez E, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.A new disease of Brachiaria was observed in 2009 at the CIAT experiment station in Palmira, Colombia, on plants of B. humidicola (CIAT accession no.16888). In 2016, the disease was observed on multiple genotypes of B. humidicola, Brachiaria hybrid cv. Mulato II, and Brachiaria hybrid Cayman. Symptoms included chlorosis along the midribs and yellowing on flag-leaf margins, followed by wilting and necrosis of foliage. Bacteria isolated from the lesions were cream-colored and produced a yellow, diffusible, non-fluorescent pigment on King´s medium B. Thirty-two bacterial strains fitting this description were pathogenic on Brachiaria spp. and were identified as Burkholderia glumae by PCR and sequence analysis of 16S rDNA. Real-time PCR was the most sensitive and accurate method evaluated for identifying the pathogen. B. glumae 88b, a highly-virulent strain identified in this study, was inoculated to ten Brachiaria genotypes including B. decumbens, B. brizantha ‘Marandú, B. brizantha ‘Toledo’, B. ruziziensis, B. brizantha ‘Piata’ and Brachiaria hybrids CIAT 36061, CIAT 36062, CIAT 36087, BR02/1752, and BR02/1794. B. glumae 88b was pathogenic on nine of the Brachiaria genotypes; interestingly, CIAT 36062 was resistant to strain 88b. This knowledge of B. glumae would help to develop bacterial blight disease management.
Introduction
Forage grasses in the genus Brachiaria are widely sown in cattle-producing regions in tropical Latin America [1,2] and are used by small-scale livestock producers in Africa and southeast Asia [3-5]. The most important commercial species include B. brizantha, B. decumbens, B. ruziziensis, and B. humidicola; these forage grasses are native to East Africa and were introduced to Latin America during the mid-20th century [6]. Since then, Brachiaria spp. have been planted on 99 million ha in Brazil alone [1], and their cultivation has drastically improved the efficiency of cattle production in areas with marginal or acid soils [2]. A recent study examined the economic impact of new Brachiaria cultivars in Brazil, Colombia and Central American countries and estimated their value at $6.7 billion [7].
The bacterial genus Burkholderia consists of more than 40 different gram-negative species that occupy a wide array of ecological niches. Burkholderia spp. occur in soil, water, fungal mycelia, the plant rhizosphere, and as endophytes in roots and shoots [8]. B. glumae (formerly Pseudomonas glumae), the causal agent of Bacterial Panicle Blight (BPB) in rice, causes economic losses in Korea, Japan, the southeastern United States and South and Central America, Africa, Vietnam, the Philippines, Taiwan, and other rice-growing areas worldwide [9-14]. Yield losses of 15% are common and losses up to 80% have been recorded in severely-infected rice fields [15]. B. glumae also cause seedling rot in nurseries [16] and has been identified as a causal agent of bacterial wilt in tomato, pepper, eggplant, sunflower and sesame [17]. B. glumae motility appears to contribute to pathogenesis as motility-deficient mutants have low virulence when inoculated in rice at the flowering stage. B. glumae is more prevalent and virulent than either B. gladioli or B. plantarii [18,19].
In Colombia, B. glumae was first reported to cause BPB in rice in 1987 [20]. Since then, the disease has been documented in many other rice-producing countries in Central and South America including Costa Rica, Cuba, the Dominican Republic, Ecuador, Nicaragua, Panama and Venezuela [21-24]. B. glumae has not been previously reported in association with Brachiaria or other tropical forage grasses; however, the temperate forage grass Lolium multiflorum (Italian ryegrass) is a known alternate host for the pathogen [25]. B. humidicola is a known host for other Burkholderia spp. including B. pseudomallei, the causal pathogen of melioidosis in humans and animals in Northern Australia [26].
In 2009, a new disease of Brachiaria was observed on B. humidicola CIAT accession no. 16888 at the CIAT experiment station in Palmira, Colombia. The disease was observed again in 2016 on B. humidicola and Brachiaria hybrids. Symptoms included a chlorotic streak along the midribs and yellowing on flag-leaf margins, followed by wilting and necrosis of leaf tissue. Initial tests indicated that B. glumae was present in the foliage of the diseased plants. In the present study, we confirmed the pathogenicity of B. glumae in Brachiaria spp. Strains of B. glumae were identified using species-specific PCR primers and subsequent sequencing of 16S rDNA. This article constitutes a report of B. glumae as the causal agent of bacterial blight of Brachiaria spp.
Materials and methods
This study was conducted in the laboratories and greenhouses of the Forage Pathology Program at CIAT, Palmira, Colombia (1001 masl; 3.5833 °N, 76.2500 °W).
Isolation and purification of bacterial strains
Bacterial strains were isolated from diseased plants (n = 120) of B. humidicola, Brachiaria hybrid cv. Mulato and Brachiaria hybrid Cayman. Brachiaria samples were collected in Palmira, Valle, in 2009 and 2016 (Table 1). The isolation of plant pathogenic bacteria was done by direct plating of infected leaf samples [27]. Small leaf fragments were washed under running tap water for 30 min, air dried on filter papers, and placed onto Nutrient Agar (NA) medium with the inclusion of a 5 min rinse in sodium hypochlorite (2% NaClO), used to sterilize the surface. Bacterial colonies were selected randomly from the disease samples for a detailed study. They included colonies with white growth typical of B. glumae and other commonly occurring colonies with cream-white growth. Isolates were originally isolated onto NA, transferred to King B and Miyajima’s medium (KB), maintained on 50% glycerol and kept in storage at -20 oC before testing. The isolates were observed under Near Ultra Violet (NUV) light (under a UV lamp) and a loopful of non-fluorescing bacterial colonies were transferred to Miyajima's medium and incubated for 2 days at 27 °C. Pure bacterial isolates from the inoculated seedlings were identified based on colony morphology on NA, pigment production on KB, and non-fluorescence of bacterial colonies.
On KB medium bacterial isolates were recovered from B. humidicola CIAT accession no.16888. These had a biochemical reaction similar to B. glumae reference isolate 3252-8. Isolated colonies on the KB medium closely resembled B. glumae colonies. They produced a non-fluorescent yellowish-green pigment.
Bacterial strains were initially stored on filter paper in darkness at -20 oC as described previously [28]. Strains were then purified by serial dilution with sterile distilled water and plated on King’s B agar (KB) (20 g protease peptone, 1.5 g KH2PO4, 1.5 g MgSO4 .7H2O, 15 g Bacto-agar and 15ml glycerol). Plates were incubated at 30 oC for 48 h. Bacterial colonies were examined for their morphological characteristics and compared to the type strain, B. glumae 3252-8. Single colonies were stored in microcentrifuge tubes at -20 oC in 50% glycerol for further characterization.
Strains of Burkholderia spp. from the American Type Culture Collection (ATCC) were used in this study and included: B. glumae (ATCC 33617), B. gladioli (ATCC 33664 and 19302), B. plantarii (ATCC 43733 and 51545), B. multivorans (ATCC BAA-247) and B. cepacia (ATCC 10856).
Pathogenicity tests
The pathogenicity of 32 putative B. glumae strains was evaluated by inoculating leaves of the interspecific Brachiaria hybrid cv. ‘Mulato II’ (CIAT 36087). Before pathogenicity tests, bacterial strains were inoculated to KB, incubated at 30 oC for 24 h, harvested with a glass rod and suspended in sterile distilled water. For inoculation, 0.1 ml of bacterial suspension (108 CFU/ml) was applied by puncturing on the surface of leaf sheaths. Lesion lengths on the inoculated sheaths were recorded 7 d after inoculation. Three replicate tests were performed for each bacterial strain and bacteria-free suspensions were used as controls.
Koch's postulates were accomplished. Firstable the bacterium was found in diseased but not healthy plants, secondly the bacterium was cultured from the diseased plants inoculated with the bacteria showing disease symptoms, and finally the bacterium was reisolated from the inoculated diseased plants and was identical to the original bacterium.
Virulence studies
The virulence of B. glumae strains was evaluated by inoculating 10 Brachiaria genotypes from the closely-related cultivated species, B. brizantha, B. decumbens, and B. ruziziensis, and hybrid cultivars derived from interspecific crosses of these species. The Brachiaria genotypes included in these tests included B. brizantha cv. ‘Marandú’ (CIAT 6294), B. brizantha cv. ‘Toledo’ (CIAT 16320), B. brizantha cv. ‘Piata’ (CIAT 16125), B. decumbens cv. ‘Basilisk’ (CIAT 606), B. ruziziensis (BRX 4402), Brachiaria interspecific hybrid CIAT 36062 and interspecific hybrid Brachiaria cultivars ‘Mulato I’ (CIAT 36061), Mulato II (CIAT 36087), ‘Cayman’ (BR02/1752) and ‘Cobra’ (BR02/1794) (Table 2). Plants from each genotype were prepared for virulence tests by detaching tillers (20 cm to 25 cm) from parental plants; tillers were then immersed for 5 min in a 1% solution of sodium hypochlorite. The tillers (clones) were then planted individually in PVC tubes (5.3 cm × 6.5 cm) containing 36 g of sterilized soil (nursery mixture of soil and sand in a 4:1 ratio). The PVC tubes and plants were arranged in a completely randomized experimental design with eight replicates and two negative controls per genotype. The transplants were grown in the greenhouse, watered every two days and fertilized one week before inoculation.
Forty-day-old clonal transplants were inoculated with 0.5 ml of a bacterial suspension (108 CFU/ml). Negative controls were inoculated with sterile distilled water. Individual inoculated plants and controls were incubated in the greenhouse at 32 °C ± 3. The above-ground portion of each plant was enclosed in a 600 ml clear plastic water bottle. The bottle acted as a micro-chamber to create conditions of high relative humidity (80% to 100%). The bottles also physically isolated each plant, preventing leaf contact between adjacent plants [29]. Each plant was evaluated for the presence or absence of symptoms (+/-) and the percentage of the total area affected 10 days after inoculation. Virulence assessments were analyzed using SAS Software v. 9 of the SAS System for Unix (Cary, North Carolina, USA) [30] using the MIXED procedure and REML estimation method.
Species-specific PCR with 16S rDNA primers
Bacterial strains (Table 1) were inoculated to KBA and incubated for 48 h at 28 oC. Bacterial cells were collected from 1.5 ml of culture (108 CFU/ml) by centrifugation at 11,000 rpm for 1 min and used for DNA isolation. Bacterial DNA was isolated using the Wizard® Genomic DNA Purification kit (Promega, Madison, WI) following the manufacturer’s instructions. The quality of DNA was checked on agarose gels and quantified using a Thermo Scientific NanoDrop 2000c UV-Vis spectrophotometer (NanoDrop, Wilmington, DE).
For identification of B. glumae, template DNA was used in PCR and amplified with 16S rDNA primers specific for B. glumae: 27f (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492r (5′-AAGGAGGTGATCCAGCC-3′), which were previously reported by Frank, et al. [31]. To identify pathogenic B. glumae strains, primers described by Sayler, et al. [32] were used in the PCR reactions: gluF (5′-ACGTTCAGGGATRCTGAGCAG-3′) and gluR (5′-AGTCTGTCTCGCTCTCCCGA-3′). Each 25 μl PCR reaction contained the following: 1 µl each of forward and reverse primers (10 ng/µl), 12.5 µl Taq polymerase (GoTaq®, Promega) at 5 U/µl, 3 µl distilled water, and 3 µl bacterial DNA at 10 ng/µl. The reactions were carried out in a thermal cycler (PTC-100 Peltier Thermal Cycler; MJ Research, Inc.) using the following conditions: initial denaturation at 94 °C for 2 min, 30 cycles of 94 °C for 1 min, 55 °C for 1 min, and 72 °C for 1 min, and final extension at 72 °C for 7 min. PCR products were resolved in 1.2% agarose gels in TAE buffer (1× Tris base, acetic acid and EDTA) and visualized with GelRed Nucleic Acid Gel Stain, 10,000 × (Life Technologies, India, Pvt. Ltd.).
Sequence analysis of 16S rRNA
Before sequence analysis, a 1500-bp PCR fragment encoding 16S rRNA was obtained from B. glumae strains with primers 27 f (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492r (5′-AAGGAGGTGATCCAGCC-3′) [31]. PCR products were purified with Polyethylene Glycol (PEG) as follows: PCR products were added to 1.5 mL of 20% PEG solution (10 g PEG 8000 and 7.3 g NaCl), mixed by vortexing, and incubated at ambient temperature for 15 min. The solution was centrifuged at 15,800 x g for 15 min at room temperature, the supernatant was discarded and 100 μl of 70% ethanol was added. This new solution was centrifuged at 15,800 x g for 2 min at ambient temperature, the supernatant was discarded, and the remaining pellet was incubated at 37 °C to dryness. The resulting DNA pellets were re-suspended in 20 μl of sterile distilled water.
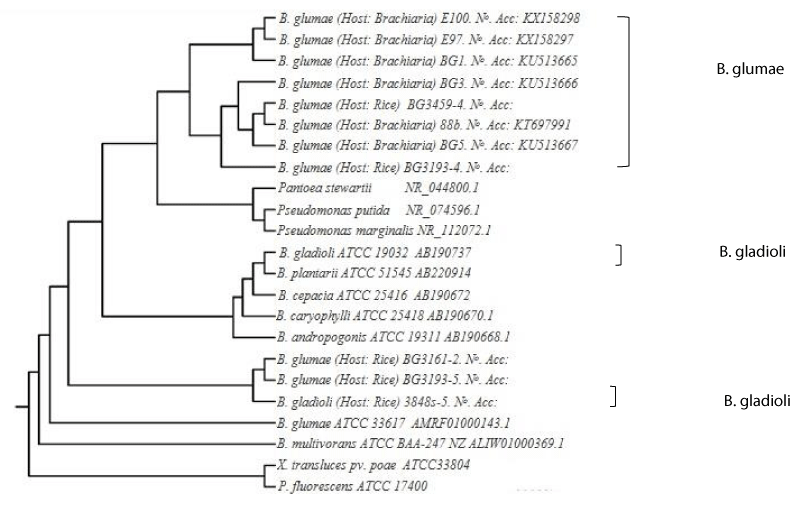
Purified PCR products were sequenced at the DNA Facility located at Iowa State University, Office of Biotechnology. 16S rDNA sequences of Burkholderia spp. from the American Type Culture Collection (ATCC) were also included for comparative purposes. These included B. glumae ATCC 33617, (GenBank accession no. AMRF01000143.1), B. gladioli ATCC 19302 (accession no. AB190737) and 51545 (accession no. AB220914), B. multivorans ATCC BAA-247 (NZ ALIW01000369.1), B. cepacia ATCC 10856, B. caryophylii ATCC 25418 (AB190670.1), B. andropogonis ATCC 19311 (AB190668.1) 16S rDNA sequences were also obtained from Pantoea stewartii ATCC 8199 (accession no. NR-044800.1), Pseudomonas putida KT2400 (NR-074596.1), Pseudomonas marginalis ATCC 10844 (NR-112072.1), Pseudomonas fluorescens ATCC 17400 (NZ JENC01000059.1) and Xanthomonas translucens pv. poae. ATCC 33804 (MADN01000503.1). B. glumae rDNA sequences from rice strains 3161-2 and 3193-5 and B. gladioli 3845s-5 (also from rice; accession no. JF 323581) were also analyzed.
The sequences were initially aligned with Chromas PRO (Technelysium Pty Ltd.) and then compared with available sequences from the European Molecular Biology Library (EMBL) and GenBank using BLASTN. Cluster analysis was performed on individual sequences of the 23 strains of our bacterial collection. Strain ATCC33804 and ATCC17400 were used to root. Using unweighted pair-group method of averages (UPGMA). This was analyzed using NTSYS PC 2.2 program and a tree was generated
Real-time PCR identification of bacterial strains
DNA was isolated for real-time PCR from B. glumae strain 3252-8 (type strain), 32 strains of B. glumae (this study), B. gladioli (rice strain no. 3845-5), Pseudomonas putida, P. marginalis, Pantoea stewartii, P. aeruginosa and Xanthomonas campestris pv. graminis. Bacterial cells (OD600 = 0.5) from 1 mL of culture were centrifuged at 8600 x g for 3 min, and the pellet was used for DNA isolation as described previously [11]. The method used for DNA isolation was also used for direct PCR and sequence analysis. Primers for real-time PCR were synthesized based on the sequence of the Internal Transcribed Spacer (ITS) of B. glumae, and primers were used to previously detect this pathogen [11]. The following primers were used for real-time PCR reactions: forward primer, FP-BUR (5′-CAAGATGATTCGAACGCAAGTT-3′); reverse primer: RP-BUR (5′-TCGCTCTCCCGAAGAGAT GA-3′); and probe P-BUR (5′-TACGGCACA AATGCGAGAACTCAACCT-3′). The real-time PCR experiment was standardized as described previously [11]. Briefly, different probe concentrations (50 to 300 ηM), primer concentrations (2.5, 5, 10, 15 and 20 pmole), and annealing and extension times (20, 30, 40 and 60 s) were tested using a uniform template concentration of B. glumae 3252-8 (type strain). The probe P-BUR (5′-TACGGCACA AATGCGAGAACTCAACCT-3′) was used for real-time reactions. The TaqMan probe was labeled with 6-carboxyfluorescein (FAM) at the 5’ end and 6-carboxy-tetramethyl-rhodamine (TAMRA) at the 3’ end as described previously [11].
Real-time PCR was performed using TaqMan universal PCR master mix and the MJ Research Thermal Cycler (PTC-100). The 30-µl reaction mixture consisted of 15 µl of 2 × master mix, 1 µl each of forward and reverse primers (5 pmol/µl), 1 µl of probe (100 ηM) and varying amounts of DNA template and sterile water. PCR conditions consisted of a 10-min preincubation period with a 15-s denaturation at 95 °C leading to 40 cycles of annealing and extension at 60 °C for 20 s. In all experiments, the nontemplate DNA control was maintained as a negative control, and three replicates were performed for each sample.
The obtained sequences were initially aligned with the Chromas PRO program and subsequently compared with sequences available in EMBL/GenBank database using BLASTn.
The dendogram was generated using the unweighted per-group method of averages (UPGMA) analysis of the DNA sequences. The NTSYS PC Version 2.2 program by pair-wise comparison was used, and the results demonstrated the overall similarities among strains (Figure 1).
Results
Brachiaria spp. grown in CIAT fields exhibited bacterial blight symptoms (Figure 2A). Bacteria were isolated from diseased Brachiaria spp. on King’s B medium and purified by serial dilution; single colonies were transferred to fresh medium. Pure cultures were stored in King’s B broth (KBB). A total of 82 bacterial strains were isolated and purified on King’s B agar medium. In this work, 32 strains (Table 1) were selected because they resembled B. glumae according to morphological characteristics (Gram-negative rods) and yellow pigmentation on KBA.
Pathogenicity tests
Bacterial strains (n = 82) from diseased Brachiaria were tested for pathogenicity on the interspecific hybrid Brachiaria cv. Mulato II. Koch’s postulates were confirmed by re-isolating B. glumae from diseased sheaths showing blight symptoms. Thirty-nine bacterial strains were pathogenic and produced a yellow pigment on KBA; 32 of these strains (Table 1) were selected for further study because they resembled B. glumae according to morphological characteristics (Gram-negative rods) and pigmentation. Ten bacterial strains were nonpathogenic and did not produce a yellow pigment on KBA.
Pathogenicity tests revealed that nine (28.2%) of the 32 tested strains were highly pathogenic in the leaves of Brachiaria cv. CIAT 36087 Mulato II. The nine highly pathogenic strains induced symptoms similar to known strains of B. glumae and included E84, E85, E91, E92, E94, E97, E100, E103 and 88b. Negative controls were nonpathogenic. All pathogenicity experiments were repeated twice.
Our results indicate that B. glumae was the pathogen causing bacterial blight in Colombia.
Virulence studies
B. glumae strain 88b exhibited virulence on Brachiaria hybrid cv. Cayman (BR02/1794), B. brizantha cv. Marandú (CIAT 6294), Brachiaria hybrid cv. Mulato II (CIAT 36087), and B. ruzizienzis BRX 4402 (Table 2). Symptoms included yellowing and chlorotic streaks in the flag leaf (Figure 2B). The percentage of leaf areas showing symptoms ranged from 15.5 to 18.0% (Table 3). Strain 88b infected all Brachiaria genotypes except Brachiaria interspecific hybrid CIAT 36062 (Table 2), which was identified as highly resistant.
Identification of strains by PCR and sequencing
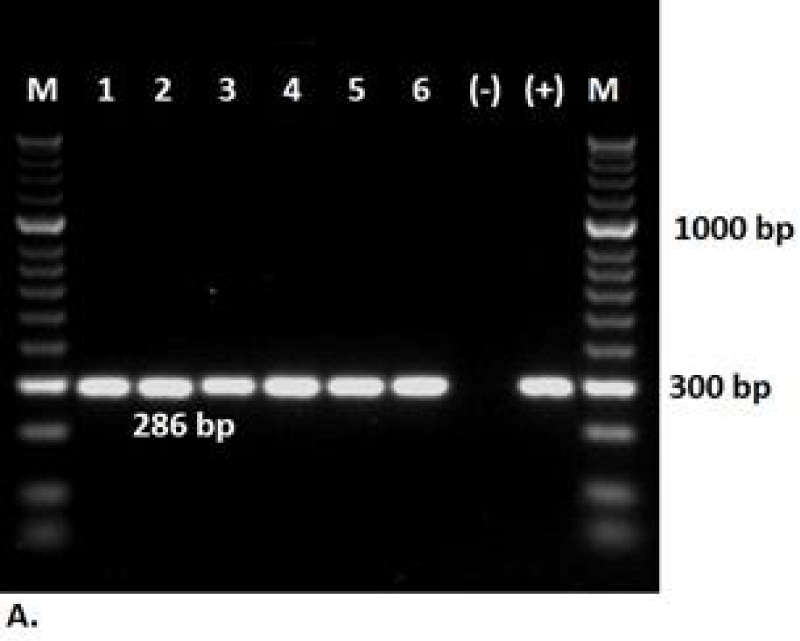
The 32 pathogenic bacterial strains were further analyzed using species-specific primers for B. glumae [32]. B. glumae strain 3252-8 was used as a positive control. Amplicons of the expected size (286 bp) were generated from six strains (Figure 3).
In the amplified region, 16S rDNA of DNA isolates from diseased Brachiaria leaves and commercial seed samples showed a 1500 bp molecular weight band, and this region was sequenced. Six bacterial strains belonging to Burkholderia glumae (GenBank accession numbers KT697991, KU513665, KU513666, KU513667, KX158297 and KX158298) were identified by PCR.
A total of 23 strains including other Burkholderia species and related species from Brachiaria and rice in Colombia were screened. Sequence analysis of the 16Sr. DNA genes from B. glumae strains (88b, BG1, BG3, BG5, E97, E100, BG 3193-4) three B. gladioli strains (B. gladioli., 3161-2, 3193- 5) showed 99% sequence similarity with already published B. glumae sequence 16SrDNA and ITS sequences for strains of the bacterial species were submitted to GenBank (accession numbers KT697991, KU513665, KU513666, KU513667, KX150277, KX158298).
By DNA sequencing, the bacterial strains were identified as Burkholderia glumae, with a sequence homology of 99% when compared with reference strains (Table 3).
Real-time PCR identification of bacterial strains
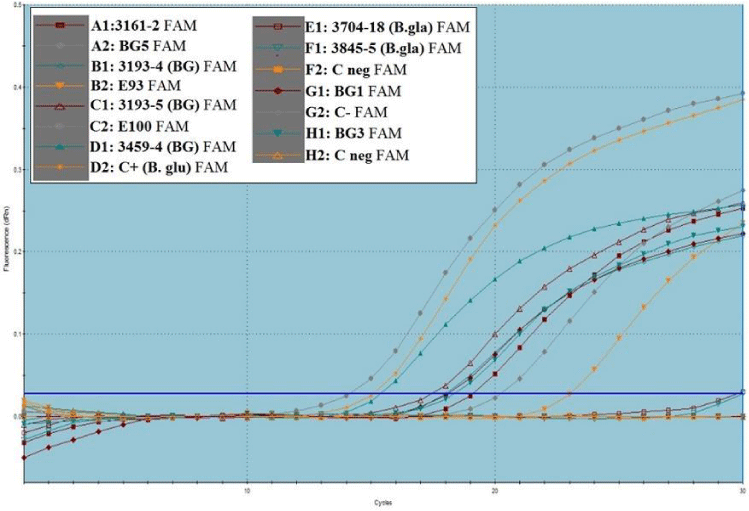
B. glumae primers FP-BUR/RP-BUR and the probe P-BUR were previously used in real-time PCR to specifically detect and quantify B. glumae [11]. Thirty amplification cycles were sufficient to obtain positive Ct values in the range of 11-16 cycles of amplification. Cycle threshold (Ct) values were compared with a standard curve to identify and determine bacterial species from tissue samples. The parameters described by Nandakumar, et al. [11] were used to develop the standard curve. Concerning the control (no template added), fluorescence was either absent or marginal until the last amplification cycle.
The specificity of primers and probe were evaluated in real-time PCR. DNA isolated from the reference strain B. glumae BG 3193-4 and 88b, BG1, BG3, BG5, E97, and E100 from Brachiaria, B. glumae strain from Colombia, and others were tested. Amplification showed that Brachiaria strains had the same amplification profile as the control strain BG 3193-4 from rice no signal was detected for B. gladioli and nontemplate control after 30 cycles
Twenty-eight strains were positively amplified in Figure 4.
These virulent, yellow-pigmented strains were previously amplified and sequenced by conventional PCR DNA sequencing and were submitted to GenBank with the following accession numbers 88b: KT697991, BG1: KU513665, BG3: KU513666, BG5: KU513667, E97: KX158297 and E100: KX158298.
Discussion
Bacterial blight symptoms have been documented in Brachiaria, but the causal pathogen was previously identified as a species of Xanthomonas [33].
This study confirmed that Burkholderia glumae isolated from Brachiaria humidicola CIAT 16888, CIAT 16889, CIAT 16890, CIAT 16891, Brachiaria hybrid CIAT 36061 cv. Mulato, and Brachiaria hybrid CIAT 36087 cv. Mulato II at CIAT (Palmira, Colombia) was pathogenic on several species of Brachiaria.
Burkholderia glumae causes both rice seedling and grain rot in the field. The bacterium produces a host-non-specific phytotoxin, toxoflavin, which is a major virulence factor [34]. Toxoflavin is a phytotoxin with a broad host range, which is a key virulence factor in bacterial grain rot. We isolated bacterial strains from leaves of Brachiaria in Colombia in 2009 and 2016. Initial observations of bacterial strains in the KBA medium and pathogenicity tests (phenotypic characteristics) suggested that B. glumae species might be involved, considering that the bacteria produce toxoflavin, resulting in yellow pigmentation on the KBA medium. By PCR and sequencing, we could determine the identity of the pathogenic strains of B. glumae with a percentage of homology of 99%.
In addition, molecular techniques have been designed for the identification of B. glumae based on specific DNA sequences in the spacer region between the genes coding for the 16S rDNA and 23S rDNA [35,]. PCR has been used to identify pathogens causing new diseases and even to differentiate species within the same genus [34,36,37]. Genotypic differences between B. glumae and B. gladioli strains were identified [18]. PCR has also been used as a method for identifying several Burkholderia species from soil, water, and infected plants [22,36-38,39]. A real-time PCR method that is effective in detecting and identifying B. glumae in seed lots and plant tissue has been used [32]. The real-time PCR assay described here detects the pathogen in leaves, eliminating the need for visualization of PCR products on an agarose gel. Real-time PCR also reduces assay time from 6h to 2h and real-time data will be available. It was the most sensitive and accurate method evaluated for identifying the pathogen.
In our pathogenicity test, the strains showed differential reactions to the pathogen. The strain 88b (B. glumae) was more virulent in genotypes B. ruziziensis (BRX 4402), B. brizantha ‘Marandú’ (CIAT 6294), Brachiaria hybrid ‘Mulato II’ (CIAT 36087), and Brachiaria hybrid BR02/1794 (Table 3).
The methodology for in vitro inoculation of leaf blades to assess pathogenicity and the microchambers methodology used in evaluating the virulence of B. glumae in genotypes of Brachiaria spp. were shown to be reliable and reproducible, allowing us to obtain reliable results concerning pathogenicity and virulence.
Burkholderia glumae was pathogenic on Brachiaria. Phytopathogenic species of Burkholderia synthesize a variety of toxins such as toxoflavin, fervenulin, and tropolone [34]. Toxoflavin is necessary for pathogenicity by Burkholderia causing both seedling disease and grain rot, for example, in rice. Several virulence factors are reported in B. glumae including catalase, flagellar biogenesis, a type III secretion system, and the main mediator of damage, toxoflavin.
The damage caused by B. glumae depends on its ability to multiply abundantly and generate high-density toxoflavin production. This causes H2O2 generation by tissue damage and obstruction of the vascular bundles on the ground, preventing the arrival of monosaccharides and disaccharides essential for the formation of foliage [10]. This obstruction also induces symptoms of chlorosis, which is responsible for bacterial wilt.
The range of interactions between these genera of bacteria and their hosts is complex and diverse. The interactions of some species seem restricted to one type of host, whereas others have a much wider host range for B. glumae. The type of interaction may be that of a pathogen, but it can also be symbiotic or both [11,17,40-42]. It is proposed that the pathogen B. glumae is the causal agent of bacterial blight in Brachiaria, showing different degrees of aggressiveness and virulence. We will therefore need to evaluate additional Brachiaria spp. genotypes against bacterial strains of B. glumae.
It has been established that the bacteria are found in the phylloplanes of rice plants [43,44], stored rice seeds [45], weeds in the field, and tissues of rice crops that were placed on the ground [46]. The genus Brachiaria helps improve soil compaction because it has roots that can penetrate the compacted subsoil. Sustainable strategies for managing bacterial blight in rice have been described [47]. Crop rotation alters the biological, chemical, and physical properties of soils, and involves planting different crops in sequence on the ground. Farmers should alternate crops to break the life cycle of pests and diseases and use various methods of control. The affected crop or crops need to be removed during these alternate periods to break the life cycle of diseases and pests that want to remain, which is an important point considering the rotation of crops involving both the host (Brachiaria) and the pathogen (B. glumae) [48]. To combat diseases, this practice is effective if the crops in the sequence are not the same and the host pathogens do not have long-term survival mechanisms in the absence of the host. Comparative genome analysis of rice-pathogenic Burkholderia provides insight into the capacity to adapt to different hosts [49]. Evaluation of major rice cultivars for resistance to bacterial seedling rot caused by B. glumae has been conducted and identification of Japanese standard cultivars for resistance assessments [50].
Screening for disease resistance using stem injection inoculation of Brachiaria grown in the greenhouse allows us to identify resistance to B. glumae. This greenhouse assay could be useful to breeders for developing cultivars resistant to bacterial blight.
This study established the genetic similarity of B. glumae strains from Colombia and the first characterization and also presents the implementation of a real-time PCR detection assay for B. glumae in Brachiaria leaves (Table 4).
Funding
International Center for Tropical Agriculture. Alvarez, E., Latorre, M. 2023. Bacterial Blight of Brachiaria caused by Burkholderia glumae in Colombia.
- Jank L, Barrios SC, Valle CB, Simeao RM, Alves GF. The value of improved pastures to Brazilian beef production. Crop Pasture Sci. 2014; 65: 1132-1137.
- Miles JW, Valle CB, Rao IM, Euclides VPB. Brachiaria grasses. 2004; 745–783 in Warm-season (C4) grasses Agron Monogr. 45, edited by LE Moser, BL Burson, LE Sollenberger. ASA, CSSA, SSSA.
- Hare MD, Phengphet S, Songsiri T, Sutin N, Vernon ESF. Impact of tropical forage seed development in villages in Thailand and Laos: Research to village farmer production to seed export. Trop. Grassl.-Forrajes Trop. 2013;1: 207–212.
- Khan ZR, Midega CA, Pittchar JO, Murage AW, Birkett MA, Bruce TJ, Pickett JA. Achieving food security for one million sub-Saharan African poor through push-pull innovation by 2020. Philos Trans R Soc Lond B Biol Sci. 2014 Feb 17;369(1639):20120284. doi: 10.1098/rstb.2012.0284. PMID: 24535391; PMCID: PMC3928888.
- Maass BL, Midega CO, Mutimura M, Rahetlah VB, Salgado P. Homecoming of Brachiaria : Improved hybrids prove useful for African animal agriculture. 2015; 8325: 27–30.
- Renvoize SA, Clayton WD, Kabuye CHS. Morphology, taxonomy and natural distribution of Brachiaria (Trin.) Griseb. 1996; 1–15 in Brachiaria: Biology, Agronomy, and Improvement, edited by JW Miles, CB Valle, BL Maass. CIAT, Palmira, Colombia.
- CIAT.The impacts of CIAT’s collaborative research. Centro Internacional de Agricultura Tropical (CIAT), Cali, Colombia. 2013.
- Vandamme P, Govan J, LiPuma J. Diversity and role of Burkholderia spp. In: Burkholderia: Molecular Microbiology and Genomics. P. Vandamme and T. Coenye, eds. Horizons Bioscience. 2006.
- Chien CC, Chang YC. The susceptibility of rice plants at different growth stages and of 21 commercial rice varieties to Pseudomonas glumae. J. Agric. Res. China. 1987; 36:302-310.
- Luo J, Xie G, Li B, Lihui X. First Report of Burkholderia glumae Isolated from Symptomless Rice Seeds in China. Plant Dis. 2007 Oct;91(10):1363. doi: 10.1094/PDIS-91-10-1363B. PMID: 30780540.
- Nandakumar R, Shahjahan AKM, Yuan XL, Dickstein ER, Groth DE, Clark CA, Cartwright RD, Rush MC. Burkholderia glumae and B. gladioli Cause Bacterial Panicle Blight in Rice in the Southern United States. Plant Dis. 2009 Sep;93(9):896-905. doi: 10.1094/PDIS-93-9-0896. PMID: 30754532.`
- Rush MC, Shao QM, Zhang S, Shahjahan AK, O’Reilly K, Shih D, Groth D, Linscombe SD. Biotechnology and control of rice diseases. La. Agric. 2003; 46:20-23.
- Karki HS, Shrestha BK, Han JW, Groth DE, Barphagha IK, Rush MC, Melanson RA, Kim BS, Ham JH. Diversities in virulence, antifungal activity, pigmentation and DNA fingerprint among strains of Burkholderia glumae. PLoS One. 2012;7(9):e45376. doi: 10.1371/journal.pone.0045376. Epub 2012 Sep 18. PMID: 23028972; PMCID: PMC3445519.
- Zhou-qi C, Bo Z, Guan-lin X, Bin L, Shi-wen H. Research status and the prospect of Burkholderia glumae, the pathogen causing bacterial panicle blight. Rice Science. 2016; 23(3):111-118.
- Fang Y, Xu L, Tian W, Yan H, Yu S, Lou MM, Xie GL. Real-time fluorescence PCR method for detection of Burkholderia glumae from rice. Rice Sci. 2009; 16:157-160.
- Azegami K, Nishiyama K, Watanabe Y, Kadota I, Ohuchi A, Fukasawa C. Pseudomonas plantarii sp. Nov., the causal agent of rice seedling blight. Int. J. Syst. Bacteriol. 1987; 37:144-152.
- Jeong Y, Kim J, Kim S, Kang Y, Nagamatsu T, Hwang I. Toxoflavin Produced by Burkholderia glumae Causing Rice Grain Rot Is Responsible for Inducing Bacterial Wilt in Many Field Crops. Plant Dis. 2003 Aug;87(8):890-895. doi: 10.1094/PDIS.2003.87.8.890. PMID: 30812790.
- Fory PA, Triplett L, Ballen C, Abello JF, Duitama J, Aricapa MG, Prado GA, Correa F, Hamilton J, Leach JE, Tohme J, Mosquera GM. Comparative analysis of two emerging rice seed bacterial pathogens. Phytopathology. 2014 May;104(5):436-44. doi: 10.1094/PHYTO-07-13-0186-R. PMID: 24261408.
- Lee C, Mannaa M, Kim N, Kim J, Choi Y, Kim SH, Jung B, Lee HH, Lee J, Seo YS. Stress Tolerance and Virulence-Related Roles of Lipopolysaccharide in Burkholderia glumae. Plant Pathol J. 2019 Oct;35(5):445-458. doi: 10.5423/PPJ.OA.04.2019.0124. Epub 2019 Oct 1. PMID: 31632220; PMCID: PMC6788416.
- Zeigler RS, Alvarez E. Bacterial sheath brown rot of rice caused by Pseudomonas fuscovaginae in Latin America. Plant Dis. 1987; 71:592-597.
- Correa F, Pérez C, Saavedra E. Añublo bacterial de la panícula del arroz. Rev. Arroz. 2007; 57(468):26-32.
- Nandakumar R, Rush MC, Correa F. Association of Burkholderia glumae and B. gladioli with Panicle Blight Symptoms on Rice in Panama. Plant Dis. 2007 Jun;91(6):767. doi: 10.1094/PDIS-91-6-0767C. PMID: 30780491.
- Quesada-Gonzales A, Garcia-Santamaria F. Burkholderia glumae in the rice crop in Costa Rica. Agron. Mesoam. 2014; 25:371-381.
- Riera-Ruiz C, Vargas J, Cedeño C, Quirola P, Escobar M, Cevallos-Cevallos JM, Ratti M, Peralta EL. First Report of Burkholderia glumae Causing Bacterial Panicle Blight on Rice in Ecuador. Plant Dis. 2014 Jul;98(7):988. doi: 10.1094/PDIS-10-13-1024-PDN. PMID: 30708859.
- Miyagawa H, Ozaki K, Kimura T. Pathogenicity of Pseudomonas glumae and P. plantarii to the ears and leaves of graminaceous plants. Bull. Chugoku Natl. Agric. Exp. Sta. 1988; 3:31-43; 17.
- Kaestli M, Schmid M, Mayo M, Rothballer M, Harrington G, Richardson L, Hill A, Hill J, Tuanyok A, Keim P, Hartmann A, Currie BJ. Out of the ground: aerial and exotic habitats of the melioidosis bacterium Burkholderia pseudomallei in grasses in Australia. Environ Microbiol. 2012 Aug;14(8):2058-70. doi: 10.1111/j.1462-2920.2011.02671.x. Epub 2011 Dec 19. PMID: 22176696; PMCID: PMC3319007.
- Zeigler RS. Aricapa G, Hoyos H. Distribution of fluorescent Pseudomonas spp. Causing grain sheath discoloration of Latin America. Plant disease. 1987; 71: 896-900.
- Aricapa MG, Correa F. Almacenamiento de hongos en papel filtro. 13 Ascolfi Inf. 1994; 20:29-30.
- Alvarez E, Latorre M, Bonilla X, Miles JW. Assessing the Resistance of Brachiaria Hybrids to Pathogenic Rhizoctonia. Plant Dis. 2014 Mar;98(3):306-310. doi: 10.1094/PDIS-04-13-0405-RE. PMID: 30708412.
- SAS Institute Inc. What's New in SAS 9.0, 9.1, 9.1.2, and 9.123? SAS Institute Inc., Cary, NC. 2004.
- Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol. 2008 Apr;74(8):2461-70. doi: 10.1128/AEM.02272-07. Epub 2008 Feb 22. PMID: 18296538; PMCID: PMC2293150.
- Sayler RJ, Cartwright RD, Yang Y. Genetic Characterization and Real-Time PCR Detection of Burkholderia glumae, a Newly Emerging Bacterial Pathogen of Rice in the United States. Plant Dis. 2006 May;90(5):603-610. doi: 10.1094/PD-90-0603. PMID: 30781136.
- Valerio JR, Lapointe SL, Kelemu S, Fernandes CD, Morales FJ. Pests and diseases of Brachiaria species. 1996; 87–105 in Brachiaria: Biology, Agronomy, and Improvement, edited by JW Miles, CB Valle, BL Maass. CIAT, Palmira, Colombia.
- Sato Z, Koiso Y, Iwasaki S, Matsuda I. Toxins produced by Pseudomonas glumae. Ann. Phytopathol. Soc. Jpn. 1989; 55:356-359.
- Maeda Y, Shinohara H, Kiba A, Ohnishi K, Furuya N, Kawamura Y, Ezaki T, Vandamme P, Tsushima S, Hikichi Y. Phylogenetic study and multiplex PCR-based detection of Burkholderia plantarii, Burkholderia glumae and Burkholderia gladioli using gyrB and rpoD sequences. Int J Syst Evol Microbiol. 2006 May;56(Pt 5):1031-1038. doi: 10.1099/ijs.0.64184-0. PMID: 16627650.
- Furuya N, Ura H, Ilyama K, Matsumoto M, Takeshita M, Takanami Y. Specific oligonucleotide primers based on sequences of the 16S-23S rDNA spacer region for the detection of Burkholderia gladioli by PCR. J. Gen. Plant Pathol. 2002; 68:220-224.
- Takeuchi T, Sawada H, Suzuki F, Matsuda I. Specific detection of Burkholderia plantarii and B. glumae by PCR using primers selected from the 16S- 23S rDNA spacer regions. Ann. Phytopathol. Soc. Jpn. 1997; 63:455-462.
- Mahenthiralingam E, Urban TA, Goldberg JB. The multifarious, multireplicon Burkholderia cepacia complex. Nat Rev Microbiol. 2005 Feb;3(2):144-56. doi: 10.1038/nrmicro1085. PMID: 15643431.
- Mahenthiralingam E, Bischof J, Byrne SK, Radomski C, Davies JE, Av-Gay Y, Vandamme P. DNA-Based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J Clin Microbiol. 2000 Sep;38(9):3165-73. doi: 10.1128/JCM.38.9.3165-3173.2000. PMID: 10970351; PMCID: PMC87345.
- Coenye T, Vandamme P. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ Microbiol. 2003 Sep;5(9):719-29. doi: 10.1046/j.1462-2920.2003.00471.x. PMID: 12919407.
- LiPuma JJ. Update on Burkholderia nomenclature and resistance. Clin. Microbiol. Newsl. 2007; 29:65-69.
- Parsons JJ. Spread of African pasture grasses to the American tropics. J. Range Manage. 1972; 25:12-17.
- Matsuda I, Sato Z. Ecology of P. glumae, cause bacterial grain rot of rice, from planting to the mature stage. Ann. Phytol. Soc. Jpn. 1987; 53:122.
- Hikichi Y. Mode of action of oxolinic acid against bacterial seedling rot of rice caused by Pseudomonas glumae. I. Relationship between population dynamics of P. glumae on seedling of rice and disease severity of bacterial seedling rots of rice. Ann. Phytol. Soc. Jpn. 1993; 59:441-446.
- Uribe AF. Principales avances en investigación y desarrollo tecnológico por sistemas de producción agrícola. CORPOICA. Mercado de Duque M, Ramírez González N, Rodríguez Quijano PA, comps. Y eds. 1998; 451.
- Thibault FM, Valade E, Vidal DR. Identification and discrimination of Burkholderia pseudomallei, B. mallei, and B. thailandensis by real-time PCR targeting type III secretion system genes. J Clin Microbiol. 2004 Dec;42(12):5871-4. doi: 10.1128/JCM.42.12.5871-5874.2004. PMID: 15583328; PMCID: PMC535269.
- Zhou XG. Sustainable Strategies for Managing Bacterial Panicle Blight in Rice. In Protecting Rice Grains in the Post-Genomic Era. IntechOpen. 2019.
- Millington S, Stopes C, Woodward L. Rotational design and the limits of organic systems ‒ the stockless organic farm? In: Proc. Symp. British Crop Protection Council. Cambridge. 1990.
- Seo YS, Lim JY, Park J, Kim S, Lee HH, Cheong H, Kim SM, Moon JS, Hwang I. Comparative genome analysis of rice-pathogenic Burkholderia provides insight into capacity to adapt to different environments and hosts. BMC Genomics. 2015 May 6;16(1):349. doi: 10.1186/s12864-015-1558-5. PMID: 25943361; PMCID: PMC4422320.
- Mizobuchi R, Fukuoka S, Tsuiki C, Tsushima S, Sato H. Evaluation of major rice cultivars for resistance to bacterial seedling rot caused by Burkholderia glumae and identification of Japanese standard cultivars for resistance assessments. Breed Sci. 2020 Apr;70(2):221-230. doi: 10.1270/jsbbs.19117. Epub 2020 Mar 19. PMID: 32523404; PMCID: PMC7272248.
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.



 Save to Mendeley
Save to Mendeley
