Scientific Journal of Genetics and Gene Therapy
Characterization of the complete chloroplast genome sequence of Vitis vinifera ‘Guifeimeigui’
Li Liu, Yang Yang, Xiujie Li* and Bo Li*
Bo Li, Shandong Academy of Grape, Shandong Academy of Agricultural Sciences, Jinan, China, E-mail: sdtalibo@163.com
Cite this as
Liu L, Yang Y, Li X, Li B (2021) Characterization of the complete chloroplast genome sequence of Vitis vinifera ‘Guifeimeigui’. Scientific J Genet Gene Ther 7(1): 001-003. DOI: 10.17352/sjggt.000019Copyright License
© 2021 Liu L, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Introduction
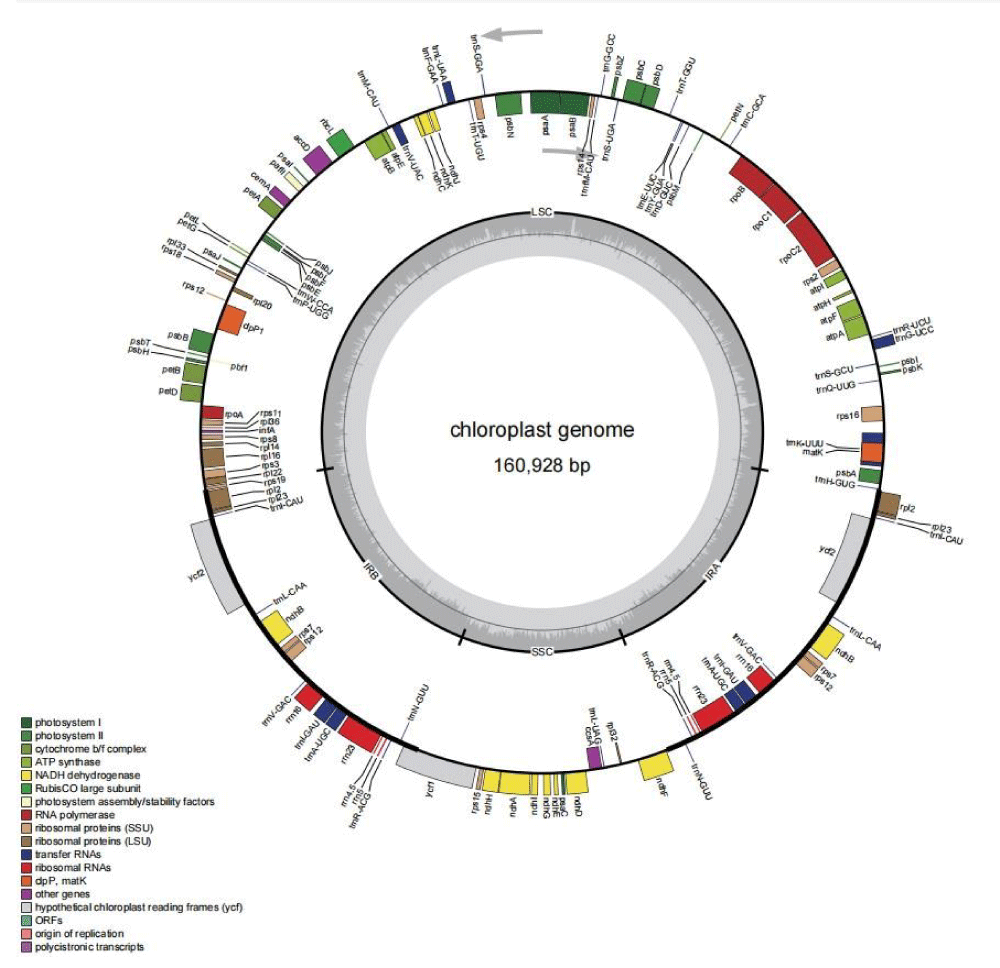
Vitis vinifera ‘Guifeimeigui’ is a diploid table grape, a Eurasian species. This research first reported the complete chloroplast (cp) genome of Vitis vinifera ‘Guifeimeigui’. The size of the complete cp genome is 160,928 bp and its GC content is 37.38%, including a pair of inverted repeats (26,353 bp each) separated by large (89,150 bp) and small (19,072 bp) single-copy regions. It encodes 85 genes, including 40 protein coding genes, 37 transfer RNA genes (tRNA), and 8 ribosomal RNA genes (rRNA). The Maximum Likelihood (ML) phylogenetic tree demonstrated that Vitis vinifera ‘Guifeimeigui’ is close to Vitis vinifera.
‘Guifeimeigui (V. vinifera) is a table grape cultivar, well known for pleasant flavors of rose, bred by a science researcher of Shandong Academy of Grape [1], Shandong Academy of Agricultural Sciences. It shows strong disease resistance and wide adaptability. It has a special rose aroma and is suitable for facilities cultivation in China. Besides, V. vinifera ‘Guifeimeigui’ is an early maturing variety. Thus, it is widely used for breeding grapes against disease and with early maturity. This high-quality assembly based on Illumina short reads sheds light on Vitis phylogeny and evolution and provides accurate genetic information for future research on Vitis species.
The sample of V. vinifera ‘Guifeimeigui’ was collected from a vineyard in Zhonggong, Jinan, China (E116°59’7”; N36°28’42”). We used fresh leaves to extract total genomic a DNA with DNeasy Plant Mini kit (Qiagen, Valencia, CA, and USA) and constructed the library with an average length of 300bp using Illumina TruSeq™ Nano DNA Sample Prep Kit. The DNA library (paired-end, PE=150bp) was sequenced on Illumina NovaSeq 6000 platform (Illumina, San Diego, CA). The resulting reads were quality-filtered using Trimmomatic v0.39 [2]. 4.38Gb clean reads were obtained in total and used to assemble the cp genome with NOVOPlasty v4.2 software [3]. The cp genome of V. acerifolia (MG664848) serves as the reference for assembly and annotation byGeSeq [3]. The complete cp genome sequence of V. vinifera ‘Guifeimeigui’ had been deposited into GenBank (No. MZ569033) and the SRA number is SRR15114330. In addition, the sample was stored at the Laboratory of Molecular Biology, Shandong Academy of Grape, Shandong Academy of Agricultural Sciences (Voucher specimen: GFMG20210416) (Li Liu, Email: 15153871569@163.com).
Chloroplast length of V. vinifera ‘Guifeimeigui’ was 160,928 bp, consisting of a large single-copy region (LSC, 89150 bp), and a small single-copy region (SSC, 19072 bp) (Figure 1). LSC and SSC are separated by a pair of inverted repeats (IRRs, 26,353 bp each). The cp genome encodes 85 genes, including 40 protein coding genes, 37 transfer RNA genes (tRNA), and 8 ribosomal RNA genes (rRNA) and 37.38% overall GC content (Figure 1). The overall GC content was very similar to that of other complete chloroplast genomes from V. vinifera [4,5]. Six protein-coding genes (rpl2, rpl23, ycf2, ndhB, rps7, rps12), seven tRNA genes (trnl-CAU, trnL-CAA, trnV-GAC, trnl-GAU, trnA-UGC, trnR-ACG, trnN-GUU) and all rRNA genes (rrn4.5, rrn5, rrn16, and rrn23) were located at the IR regions. The GC content of the LSC (35.31%) and SSC (31.65%) regions was lower than those in the IRa region (42.96%) and the IRb region (42.97%). Therefore, the relatively high GC content of the IR regions was mostly attributable to the four rRNAs and tRNAs [6,7].
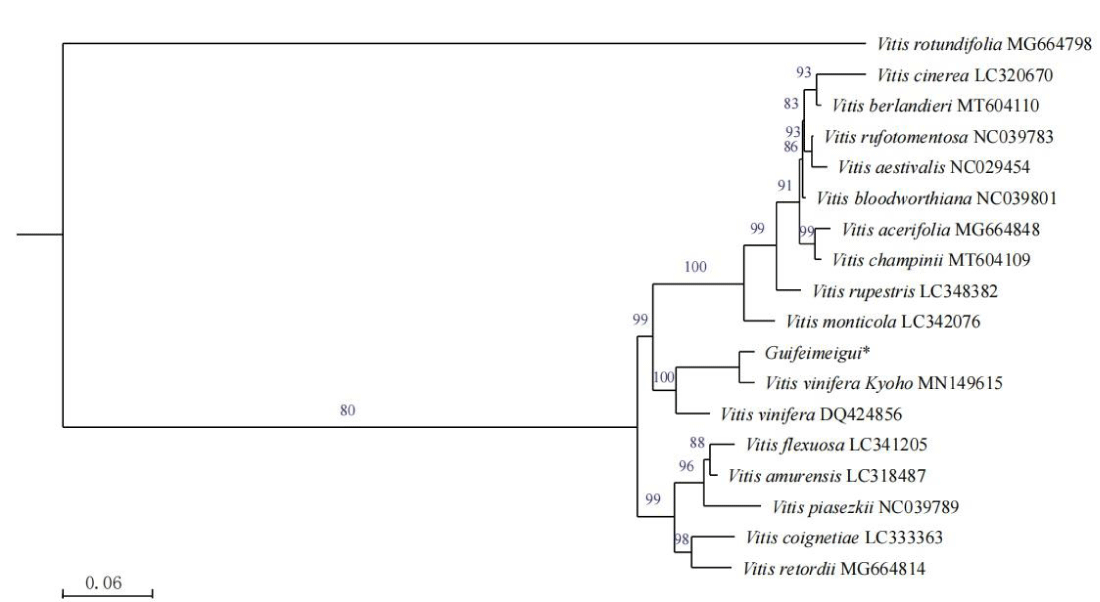
To investigate the phylogenetic position of V. vinifera ‘Guifeimeigui’ and other 17 V. species, a maximum likelihood phylogenetic tree was constructed (Figure 2) through the PhyML [8,9]. The result revealed that V. vinifera ‘Guifeimeigui’ is situated within the same branch as V. vinifera (MN149615), which suggested that ‘Guifeimeigui’ has more traits of the V. vinifera.
The authors would like to thank TopEdit (www.topeditsci.com) for linguistic assistance during the preparation of this manuscript.
Funding
This study was funded by the Natural Science Foundation of Shandong Province (Grant No. ZR201807090168), New Variety Breeding of High-Quality Characteristic Fruit Trees (Grant No. 2020LZGC008), and Integrated Research and Demonstration of Key Techniques of Facility Cultivation of High Quality and Efficiency Grape (2020ZX010).
Data availability
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession No. MZ569033, or available from the corresponding author.
- Li KY (2015) 14 grape varieties suitable for early cultivation in solar greenhouse. Gansu Agricultural Science and Technology 74-77.
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114-2120. Link: https://bit.ly/3pqqGf0
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, et al. (2017) GeSeq–Versatile and accurate annotation of organelle genomes. Nucleic Acids Res 45: W6-W11. Link: https://bit.ly/3rtmApb
- Guo DH, Li D, Li H, Wang S, Wang L, et al. (2020) The complete chloroplast genome sequence of Vitis vinifera Muscat Hamburg. Mitochondrial DNA B Resour 5: 117-118. Link: https://bit.ly/3rv76RB
- Zhang L, Guo D, Liu M, Dou F, Ren Y, et al. (2021) The complete chloroplast genome sequence of Vitis vinifera × Vitis labrusca 'Shenhua'. Mitochondrial DNA B Resour 6: 166-167. Link: https://bit.ly/3ogWjbI
- Shen X, Wu M, Liao B, Liu Z, Bai R, et al. (2017) Complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Artemisiaannua. Molecules 22: 1330. Link: https://bit.ly/3pr2Hw6
- Asaf S, Khan AL, Khan MA, Waqas M, Kang SM, et al. (2017) Chloroplast genomes of Arabidopsis halleri ssp.gemmifera and Arabidopsis lyrata ssp.petraea:Structures and comparative analysis. Scitific Repprts 7: 7556. Link: https://go.nature.com/3IcE5QF
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, et al. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of phyml 3.0. Systematic Biology 59: 307-321. Link: https://bit.ly/3ly8COZ
- Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version7.0 for bigger datasets. Mol Biol Evol 33: 1870-1874. https://bit.ly/3oiYFqx
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
