Scientific Journal of Genetics and Gene Therapy
Identification of Novel TMC1 Compound Heterozygous Mutations Related to Autosomal Recessive Hearing Loss by Targeted Capture Sequencing
Xue Gao1,2, Sha-Sha Huang1, Yu Su1, Jin-Cao Xu and Pu Dai1,2*
2Department of Otorhinolaryngology, General Hospital of the Rocket Force, 16# Xin Wai Da Jie, Beijing, 100088, PR China
Cite this as
Gao X, Huang SS, Su Y, Xu JC, Dai P (2016) Identification of Novel TMC1 Compound Heterozygous Mutations Related to Autosomal Recessive Hearing Loss by Targeted Capture Sequencing. Scientific J Genetics Gen Ther 2(1): 013-016. DOI: 10.17352/sjggt.000009Mutations in the transmembrane channel-like gene1 (TMC1) are known to cause autosomal dominant and recessive forms of nonsyndromic hearing loss DFNA36 and vDFNB7/11, respectively. Here, we characterized a 5-year old girl with severe sensorineural hearing loss. By combining targeted capture of 140 known deafness genes, next-generation sequencing and bioinformatic analysis, we identified two compound heterozygous mutations TMC1 c.1333C>T (p.R445C) and c.1765A>G (p.M589V) as the disease-causing mutations. Our results indicated that TMC1 c.1333C>T (p.R445C) and c.1765A>G (p.M589V) lead to hearing loss in this family and highlight the power of combining deafness gene capture and next generation sequencing for identification of defective genes in sporadic cases with hearing loss. This approach will facilitate identification of the mutation spectrum of all known
Background
Hearing loss is a common sensory defect in humans. Nonsyndromic hereditary forms, in which hearing loss is the only clinical sign, are known to be genetically heterogeneous [1]. To date, more than 200 genetic loci and 140 responsible genes for hereditary hearing loss have been identified (Hereditary Hearing Loss Homepage, http:// hereditaryhearingloss.org/). The extremely genetic heterogeneity of hearing impairment is a major challenge for traditional genetic testing and counseling. Targeted DNA capture and massively parallel sequencing are ideal tools to address this challenge.
Next generation sequencing, is a revolutionary technology that allows large amounts of genomic sequence information to be obtained rapidly and at a low cost [2]. Because of its large capacity to survey the whole exome and genome in an unbiased manner, NGS (next generation sequencing) is well suited to identifying the causative mutations of hereditary hearing loss. NGS has commonly been used to identify disease genes within even a limited number of patient samples [3-7]. Novel genes for non-syndromic [8,9] and syndromic [10], hearing loss have also been identified using the targeted NGS approach. Targeted deaf gene enrichment usually increases this proportion by at least 1000 fold [11,12]. In this study, we applied targeted capture of 140 known deafness genes, next-generation sequencing and bioinformatic analysis to analyze the genetic pathogenesis of one sporadic patient with severe sensorineural.
TMC1 is predicted to encode membrane proteins with six transmembrane domains that may be involved in the functional maturation of cochlear hair cells [13]. Although the specific functions of the protein encoded by TMC1 are unknown, bioinformatic analysis [14] and data obtained from in vitro heterologous systems [13], suggest that TMC1 functions as a membrane channel or transporter [15]. TMC1 mutations cause nonsyndromic autosomal dominant and recessive hearing loss at the DFNA36 and DFNB7/11 loci, respectively. More than 30 recessive mutations in TMC1 are associated with autosomal recessive nonsyndromic hearing loss (ARNSHL) at the DFNB7/11 locus in 39 families worldwide [14,16-24].
In this study, we identified one small ARNSHL pedigree with severe sensorineural hearing loss caused by the c.1333C>T (p.R445C) and c.1765A>G (p.M589V) mutation of TMC1 through targeted deaf gene capture, next generation sequencing and bioinformatic analysis. This is the first study to identify TMC1 c.1333C>T (p.R445C) and c.1765A>G (p.M589V) as the ARNSHL-associated mutation in this family.
Materials and Methods
Clinical data
Family C01701 is a two-generation Chinese family with autosomal recessive nonsyndromic hearing loss. To identify candidate mutations, DNA samples were obtained from 3 members of family C01701. Mutations of GJB2 and SLC26A4 had been excluded previously. Fully informed written consent was attained from two guardians. The study was approved by the Chinese PLA General Hospital ethics of research committees. Clinical information was gathered through multiple interviews with the family members. Medical history was obtained using a questionnaire regarding the following aspects of this condition: subjective degree of hearing loss, age at onset, evolution, symmetry of the hearing impairment, use of hearing aids, presence of tinnitus, medication, noise exposure, pathological changes in the ear and other relevant clinical manifestations. Otoscopy, physical examination and pure tone audiometric examination (at frequencies from 250 to 8000 Hz) were performed to identify the phenotype. Immittance testing was applied to evaluate middle-ear pressure, ear canal volume and tympanic membrane mobility. Unaffected phenotype status was defined by a threshold lower than age- and gender-matched 50th percentile values for all frequencies measured. Physical examination of all members revealed no signs of systemic illness or dysmorphic features. CT scans of the temporal bone in the patient were performed. The diagnosis of profound sensorineural hearing impairment was made according to the WHO criteria based on audiometric examination. Tandem gait and Rhomberg tests were performed to evaluate balance.
Deafness gene capture and Illumina library preparation
Each DNA sample is quantified by agarose gel electrophoresis and Nanodrop (Thermo). Libraries were prepared using Illumina standard protocol. Briefly, 3 microgram of genomic DNA was fragmented by nebulization, the fragmented DNA is repaired, Illumina adapters are then ligated to the fragments, and the sample is size selected aiming for a 350–400 base pair product. The size selected product is PCR amplified (Each sample is tagged with a unique index during this procedure), and the final product is validated using the Agilent Bioanalyzer.
The amplified DNA was captured with a deafness related Gene Panel using biotinylated oligo-probes (MyGenostics GenCap Enrichment technologies). The probes were designed to tile along 140 deafness related genes. The capture experiment was conducted according to manufacturer’s protocol. The enrichment libraries were sequenced on Illumina HiSeq 2000 sequencer for paired read 100bp.
Bioinformatics analysis
For sequencing analysis, high-quality reads were retrieved from raw reads by filtering out the low quality reads and adaptor sequences using the Solexa QA package and the cutadapt program (http://code. google.com/p/cutadapt/), respectively. SOAPaligner program was then used to align the clean read sequences to the human reference genome (hg19).
After the PCR duplicates were removed by the Picard software, the SNPs was firstly identified using the SOAPsnp program (http:// soap.genomics.org.cn/soapsnp.html). Subsequently, we realigned the reads to the reference genome using BWA and identified the insertions or deletions (InDels) using the GATK program (http:// www.broadinstitute.org/gsa/wiki/index.php/Home_Page). The identified SNPs and InDels were annotated using the Exome-assistant program (http://122.228.158.106/exomeassistant). Magic Viewer was used to view the short read alignment and validate the candidate SNPs and InDels. Nonsynonymous variants were evaluated by four algorithms, Ployphen, SIFT, PANTHER and Pmut, as described previously to determine pathogenicity.
Mutational analysis of TMC1
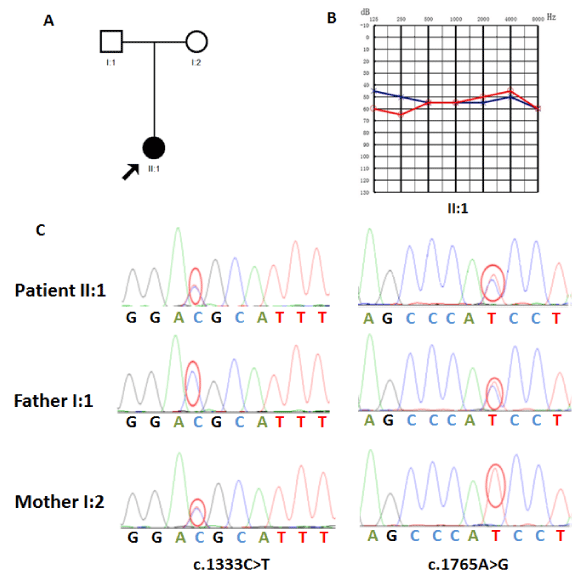
The segregation of the TMC1 c.1333C>T (p.R445C) and c.1765A>G (p.M589V) mutations was tested in 3 family members 014(I:1, I:2 and II:1), including the one whose gDNAs has been subjected to 140 deafness-associated gene NGS analysis, using PCR followed by bidirectional Sanger sequencing of the amplified fragments (ABI 3100, Applied Biosystems, USA). Nucleotide alteration(s) was identified by sequence alignment with the TMC1 GenBank sequence using the Genetool software.
Multiple sequence alignment
Multiple sequence alignment was performed according to a Homologene program with default settings and the sequences NP_619636.2 (H. sapiens), XP_001093188.2 (M. mulatta), XP_528322.2 (P. troglodytes), XP_002689695.2 (B. taurus), XP_005616877.1 (C. lupus), NP_083229.1 (M. musculus), NP_001101991.1 (R. norvegicus), NP_001006580.1 (G. gallus) and XP_695621.4 (D. rerio).
(http://www.ncbi.nlm.nih.gov/homologene?cmd=Retrieve&dopt=MultipleAlignment&list_uids=23670)
Results
Family and clinical evaluations
Family C01701 is a two-generation Chinese family with severe sensorineural hearing loss (Figure 1A) and includes 1 affected patients: II:1 (female, 5 years old). Hearing impairment was severe, prelingual and stable (Figure 1B).
Vestibular analysis was performed in the patient II:1 who did not complain about dizziness, vertigo, or imbalance. All position tests produced no nystagmus without vertigo sensation. II:1 did not have obvious delayed gross motor development. The physical examinations revealed no signs of systemic illness. High-resolution computed tomography of the temporal bone in II:1 was normal, excluding inner-ear malformations. This phenotype was consistent with that reported for DFNB7/11.
Targeted deafness gene capture and massively paralleled sequencing
We sequenced all the coding exons plus ~100 bp of the flanking intronic sequence of 140 deafness genes in one affected (II:1) member of family C01701. Under the autosomal recessive or autosomal dominant de novo mode, two variants leading to amino acid change were detected in TMC1: c.1333C>T (p.R445C) and c.1765A>G (p.M589V). Of these, one variant, c.1333C>T (p.R445C), was reported previously as a pathogenic mutation [25], whereas c.1765A>G (p.M589V) was novel and had not been found in the dbSNP databases.
Mutation detection and analysis
Using Sanger sequencing, three participating family members (1 affected and 2 unaffected) in family C01701 were genotyped to identify the mutations. Compound heterozygous c.1333C>T and c.1765A>G mutations of TMC1 were identified in the affected family member of the second generation (II:1). TMC1 p.R445C was found heterozygous in mother with normal hearing (I:2), whereas p.M589V was found heterozygous in father with normal hearing (I:1) (Figure 1C), which is consistent with autosomal recessive inheritance.
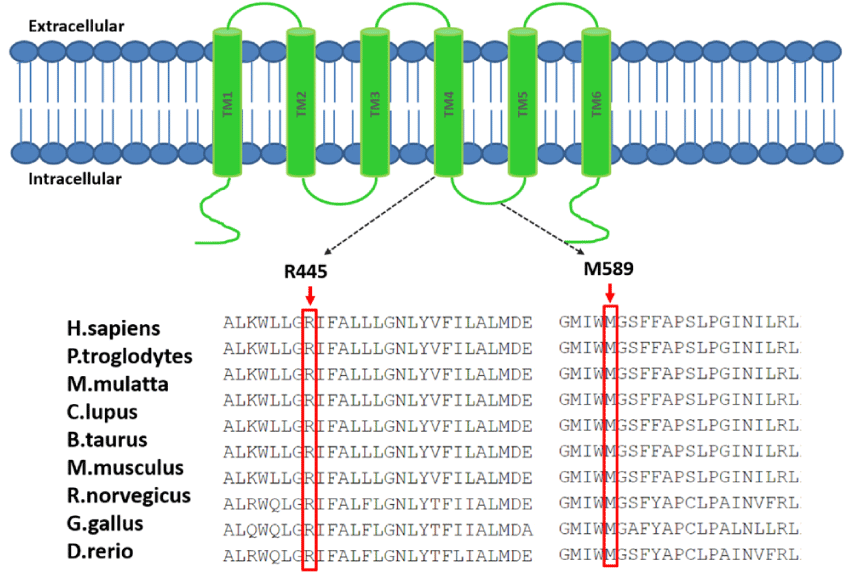
We used TMHMM2.0 to predict the TMC1 protein with six membrane-spanning regions and cytoplasmic N and C terminal regions. R445C located in the TM4 domain and p.M589V is located in the cytoplasm between the TM4 and TM5 domains of TMC1. This predicted model of TMC1 membrane topology is similar to those for ion channels and transporters.
Two amino acid substitutions occurred in revolutionarily conserved regions across different species (Figure 2). It was predicted that both TMC1c.1333C>T and c.1765A>G mutations would be damaging by SIFT and Polyphen2.
Discussion
In this study, we identified TMC1 c.1333C>T (p.R445C) and c.1765A>G (p.M589V) as the disease-causing mutation in a twogeneration Chinese family (C01701) with ARNSHL through multiple deaf genes capture, next-generation sequencing and bioinformatic analysis.
Massively parallel sequencing is a revolutionary technology that enables us to obtain large amounts of genomic sequence information in a rapid and low-cost manner [2]. With targeted gene capture, the proportion of DNA fragments containing or near targeted regions is greatly increased. Because of its ability to enrich deafness genes, NGS can be used to identify causative mutations of hereditary hearing loss. The use of NGS has frequently resulted in the identification of disease genes within a limited number of patient samples [4-7,26,27].
Mouse models with TMC1 defects [14, 15] support a role for TMC1 in the inner and outer hair cells, either in proper trafficking of other membrane proteins in these cells or in regulating the differentiation of immature hair cells into fully functional auditory receptors [13]. This model proposed that mechanical forces brought about by bending of stereocilia and tension on the tip links directly activate ion channels. If it is true that TMC1 is an ion channel localized mainly in the IHC, then it might be involved in the most basic auditory process of hair-cell transduction.
The predicted structure of TMC1 is similar to that of the α-subunit of voltage-dependent K+ channels, which has six α-helical TM segments and intracellular N and C termini [28]. It was predicted that TMC1 might be an ion channel or transporter which mediated K+ homeostasis in the inner ear [29]. The first four TM domains of the K+ channel α-subunit act as voltage sensors for activation gating [30], whereas the intervening segment between TM5 and TM6 appears to confer channel selectivity [28]. One novel conserved TMC1 sequence variant in this study c.1765A>G (p.M589V) lies within a large cytoplasmic loop between TM4 and TM5 that includes the TMC domain and a potential pore-forming loop [31].
Conclusions
Overall, we report here the clinical and genetic characteristics of one small Chinese family with ARNSHL. Identification of novel mutations has an important impact on clinical patient management, genetic counseling, molecular diagnosis, and development of advanced therapeutic strategies.
These investigations were supported by National Natural Science Foundation of China (81230020, 81371096) to P.D. Grants from China Postdoctoral Science Foundation (No. 2012M521878, No. 2013T60947) and National Natural Science Foundation of China (81570929) to X.G. Grants from National Natural Science Foundation of China (81400471) to Y.S. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We sincerely thank all the family members for their participation and cooperation in this study.
- Friedman TB, Griffith AJ (2003) Human nonsyndromic sensorineural deafness. Annu Rev Genomics Hum Genet 4: 341-402.
- Metzker ML (2010) Sequencing technologies - the next generation. Nat Rev Genet 11: 31-46.
- Simpson DA, Clark GR, Alexander S, Silvestri G, Willoughby CE (2011) Molecular diagnosis for heterogeneous genetic diseases with targeted highthroughput DNA sequencing applied to retinitis pigmentosa. J Med Genet 48: 145-151.
- Kalay E, Yigit G, Aslan Y, Brown KE, Pohl E, et al. (2011) CEP152 is a genome maintenance protein disrupted in Seckel syndrome. Nat Genet 43: 23-26.
- Krawitz PM, Schweiger MR, Rodelsperger C, Marcelis C, Kolsch U, et al. (2010) Identity-by-descent filtering of exome sequence data identifies PIGV mutations in hyperphosphatasia mental retardation syndrome. Nat Genet 42: 827-829.
- Kuhlenbaumer G, Hullmann J, Appenzeller S (2011) Novel genomic techniques open new avenues in the analysis of monogenic disorders. Hum Mutat 32: 144-151.
- Musunuru K, Pirruccello JP, Do R, Peloso GM, Guiducci C, et al. (2010) Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N Engl J Med 363: 2220-2227.
- Rehman AU, Morell RJ, Belyantseva IA, Khan SY, Boger ET, et al. (2010) Targeted capture and next-generation sequencing identifies C9orf75, encoding taperin, as the mutated gene in nonsyndromic deafness DFNB79. Am J Hum Genet 86: 378-388.
- Walsh T, Shahin H, Elkan-Miller T, Lee MK, Thornton AM, et al. (2010) Whole exome sequencing and homozygosity mapping identify mutation in the cell polarity protein GPSM2 as the cause of nonsyndromic hearing loss DFNB82. Am J Hum Genet 87: 90-94.
- Pierce SB, Walsh T, Chisholm KM, Lee MK, Thornton AM, et al. (2010) Mutations in the DBP-deficiency protein HSD17B4 cause ovarian dysgenesis, hearing loss, and ataxia of Perrault Syndrome. Am J Hum Genet 87: 282-288.
- Brownstein Z, Friedman LM, Shahin H, Oron-Karni V, Kol N, et al. (2011) Targeted genomic capture and massively parallel sequencing to identify genes for hereditary hearing loss in Middle Eastern families. Genome Biol 12: R89.
- Shearer AE, DeLuca AP, Hildebrand MS, Taylor KR, Gurrola J, et al. (2010) Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci U S A 107: 21104-21109.
- Marcotti W, Erven A, Johnson SL, Steel KP, Kros CJ (2003) Tmc1 is necessary for normal functional maturation and survival of inner and outer hair cells in the mouse cochlea. J Physiol 574: 677-698.
- Kurima K, Peters LM, Yang Y, Riazuddin S, Ahmed ZM, et al. (2002) Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat Genet 30: 277-284.
- Vreugde S, Erven A, Kros CJ, Marcotti W, Fuchs H, et al. (2002) Beethoven, a mouse model for dominant, progressive hearing loss DFNA36. Nat Genet 30: 257-258.
- Hilgert N, Alasti F, Dieltjens N, Pawlik B, Wollnik B, et al. (2008) Mutation analysis of TMC1 identifies four new mutations and suggests an additional deafness gene at loci DFNA36 and DFNB7/11. Clin Genet 74: 223-232.
- Kalay E, Karaguzel A, Caylan R, Heister A, Cremers FP, et al. (2005) Four novel TMC1 (DFNB7/DFNB11) mutations in Turkish patients with congenital autosomal recessive nonsyndromic hearing loss. Hum Mutat 26: 591.
- Kitajiri SI, McNamara R, Makishima T, Husnain T, Zafar AU, et al. (2007) Identities, frequencies and origins of TMC1 mutations causing DFNB7/B11 deafness in Pakistan. Clin Genet 72: 546-550.
- Meyer CG, Gasmelseed NM, Mergani A, Magzoub MM, Muntau B, et al. (2005) Novel TMC1 structural and splice variants associated with congenital nonsyndromic deafness in a Sudanese pedigree. Hum Mutat 25: 100.
- Santos RL, Wajid M, Khan MN, McArthur N, Pham TL, et al. (2005) Novel sequence variants in the TMC1 gene in Pakistani families with autosomal recessive hearing impairment. Hum Mutat 26: 396.
- Tlili A, Rebeh IB, Aifa-Hmani M, Dhouib H, Moalla J, et al. (2008) TMC1 but not TMC2 is responsible for autosomal recessive nonsyndromic hearing impairment in Tunisian families. Audiol Neurootol 13: 213-218.
- Gao X, Su Y, Guan LP, Yuan YY, Huang SS, et al. (2013) Novel compound heterozygous TMC1 mutations associated with autosomal recessive hearing loss in a Chinese family. PLoS One 8: e63026.
- Hassan MA, Shah AA, Szmida E, Smigiel R, Sasiadek MM, et al. (2015) A TMC1 (transmembrane channel-like 1) mutation (p.S320R) in a Polish family with hearing impairment. J Appl Genet 56: 311-316.
- Davoudi-Dehaghani E, Fallah MS, Tavakkoly-Bazzaz J, Bagherian H, Zeinali S (2015) Allelic heterogeneity among Iranian DFNB7/11 families: report of a new Iranian deaf family with TMC1 mutation identified by next-generation sequencing. Acta Otolaryngol 135: 125-129.
- Sirmaci A, Duman D, Ozturkmen-Akay H, Erbek S, Incesulu A, et al. (2009) Mutations in TMC1 contribute significantly to nonsyndromic autosomal recessive sensorineural hearing loss: a report of five novel mutations. Int J Pediatr Otorhinolaryngol 73: 699-705.
- Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, et al. (2011) Wholegenome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 475: 101-105.
- Simpson CL, Justice CM, Krishnan M, Wojciechowski R, Sung H, et al. (2011) Old lessons learned anew: family-based methods for detecting genes responsible for quantitative and qualitative traits in the Genetic Analysis Workshop 17 mini-exome sequence data. BMC Proc 5: S83.
- Hanlon MR, Wallace BA (2002) Structure and function of voltage-dependent ion channel regulatory beta subunits. Biochemistry 41: 2886-2894.
- Keresztes G, Mutai H, Heller S (2003) TMC and EVER genes belong to a larger novel family, the TMC gene family encoding transmembrane proteins. BMC Genomics 4: 24.
- Li-Smerin Y, Hackos DH, Swartz KJ (2000) alpha-helical structural elements within the voltage-sensing domains of a K(+) channel. J Gen Physiol 115: 33-50.
- Labay V, Weichert RM, Makishima T, Griffith AJ (2010) Topology of transmembrane channel-like gene 1 protein. Biochemistry 49: 8592-8598.
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
