Open Journal of Plant Science
Phylogenetic and evolutionary relationships in selected Pinus species using rbcL and matK chloroplast genes
Lav Singh1,2, Pooja Dixit1, Ravi Prakash Srivastava1, Shivaraman Pandey3, Arpit Singh2, Praveen Chandra Verma3 and Gauri Saxena1*
2Postgraduate Department of Botany, Munger University, RD and DJ College Campus, Munger -811201, India
3Plant Diversity, Systematics and Herbarium Division, CSIR-National Botanical Research Institute, Lucknow 226001, India
4Plant Molecular Biology and Genetic Engineering Division, CSIR-National Botanical Research Institute, Lucknow, 226001, India
Cite this as
Singh L, Dixit P, Srivastava RP, Pandey S, Saxena G, et al. (2021) Phylogenetic and evolutionary relationships in selected Pinus species using rbcL and matK chloroplast genes. Open J Plant Sci 6(1): 064-068. DOI: 10.17352/ojps.000035The genetic diversity of Pinus plants has been assessed in various phylogenetic studies that reveal the polymorphism directly at DNA levels. The rbcL and matK are the most commonly used markers for phylogenetic studies of Pinus sp. that exhibit a diverse geophysical adaptiveness and geographical variations across different regions as a result of genotypic modifications. This study evaluated usefulness of rbcL and matK genes for molecular identification and phylogenetic study among various species of Pinus. Maximum Likelihood (ML) and Neighbor Joining (NJ) analysis of the data obtained from rbcL gene belonging to ten Pinus species revealed four clusters. First Cluster included P. Wallichiana (PW), P.Elliottii (PEl) and P.Greggii (PG). Second cluster included P. Thunbergii(PT), P. echinata (PE) and P.merkusii (PM). Third cluster included P.taeda (Ptd) and P. Khasya (PK) while fourth cluster included P. Roxburghii(PR) and P. patula (PP). Similarly ML and NJ analysis of the data obtained from matK gene belonging to ten Pinus species revealed four cluster and one outgroup. First Cluster included P. Thunbergii(PT), P.Greggii (PG) and P.Elliottii (PEl) Second cluster included P. roxburghii(PR) and P.merkusii (PM)P. echinata (PE) and third cluster included P. Wallichiana (PW) and P.taeda (Ptd) while fourth cluster included P. Patula (PP) and P. Khasya (PK). P.echinata remained out clustered in this analysis.
Abbreviations
CTAB: Cetyl Trimethylammonium Bromide; PCR: Polymerase Chain Reaction
Introduction
Pinus (Pinaceae) is the largest genus of order Coniferales widely distributed throughout temperate zones in the Northern Hemisphere, and restricted to high elevation in the tropics and subtropics [1,2] Pines differ from other members of family Pinaceae and are easily identifiable by their dimorphic long and short shoots called fascicles. These fascicles bear long, narrow needle-like leaves mostly present in groups of two to five. Due to its great economical and ecological importance, phylogeny and systematics of this genus has received great attention based on morphology, anatomy, ethnobotanical values, karyotypic analysis, secondary products, isozymes, pharmacognosy and, most recently, molecular studies [2]. Based on morpho-anatomical and molecular data, Pinus has been divided into two monophyletic subgenera: Haploxylon (subgenus Strobuswith single fibrovascular bundle in the needle, also known as “Soft pines”) and Diploxylon (subgenus Pinus, with double fibrovascular bundles in the needle, also known as “Hard pines”) [3-6]. These two subgenera namely Haploxylon and Diploxylon have further been divided into many sections and subsections [1,4].
In this study, we analyzed sequences from rbcL and matK, for ten species of the Pinus namely P.merkusii, P.khasya, P.thunbergii, P.wallichiana, P.roxburghii, P.taeda, P.elliottii, P.echinata, P.patula and P.greggii. Out of ten species selected for our study five species viz. P.merkusii, P.khasya, P.thunbergii, P.wallichiana and P.roxburghii are native to Indian subcontinent and grow luxuriantly in Indian Himalayas while remaining five viz. P.taeda, P.elliottii, P.echinata, P.patula and P.Greggii are exotic species and have been introduced in Indian subcontinent. Our main objectives during the present study was to examine the phylogenetic relationships of selected species which are growing naturally from subgenus Pinus and Strobus at the inter- and intrasectional levels.
Materials and methods
Collection of plant material
The leaves (needles) of 10 pine species were collected in September, 2016, from a wild population in the region of Ranikhet (located at 357 km NSE of New Delhi, latitude 29°39’52.2" (N); longitude 79°28’40.9" (E); altitude 1,727 m). The site received median rainfall, had very low winter temperature and low soil fertility. Voucher specimen of all the species selected for study was deposited in the herbarium of the National Botanical Research Institute, Lucknow (Table 1). Prior to extract preparation, study material was dried at room temperature and ground to fine powder in a blender. The collected plant material was brought to the laboratory and stored at -800C.
DNA extraction, amplification and sequencing
DNA was extracted from 100 mg of dried needles using a modified CTAB method, treated with RNase, and purified by phenol. Two cpDNA regions (rbcL and matK) were sequenced using the primers (Table 2) designed by Wang, et al. [7]. PCR amplifications were accomplished at 95oC for 5 min for the initial denaturation followed by 35 cycles of denaturation at 95oC for 45 sec, annealing at 48oC (matK) and 52oC (rbcL) extension at 72oC for 2 min, and a final extension for 10 min at 72oC. The products from PCR were precipitated using ethanol and used as a template for the sequence reaction. The sequencing was carried out using an ABI 310 Genetic Analyzer (Applied Biosystems) with an ABI BigDye Terminator Cycle Reaction Kit following the manufacturer’s instructions.
Alignments were performed using Clustal X [8]. The phylogenetic analyses were completed using single data sets. The phylogenetic and evolutionary analyses were performed with MEGA X using neighborjoining (NJ) and maximum likelihood (ML) algorithms. Gel electrophoresis representing DNA bands of rbcL and matK are represented in Figures 1,2 respectively.
Results
Nucleotide variation
Composition of nucleotide from both genes was dominated of A-T (Adenine-Thymine) than GC (Guanine-Cytosine) and it was found to be A=28.76; T=26.24; C=25.07; G=19.92 and A=32.27; T=30.90; C=18.49; G=18.32 in rbcL and matK respectively (Table 2).
Phylogenetic relationships among selected species of Pinus
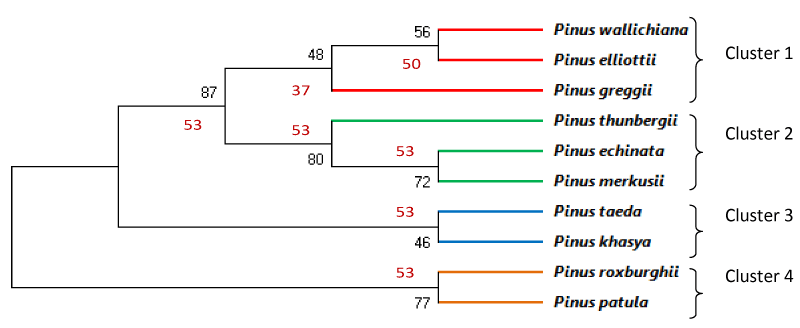
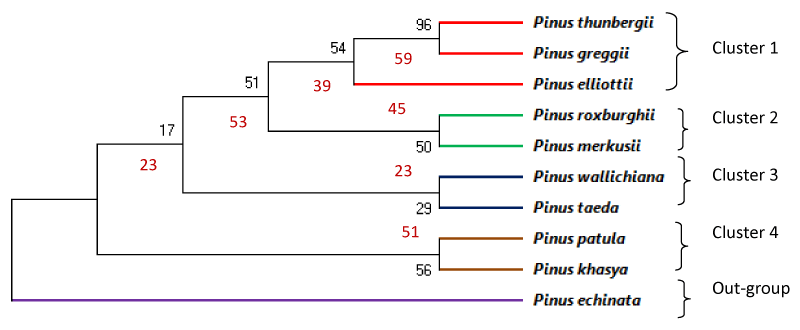
The results of the tree construction were midpoint rooting based on partial sequences of rbcL gene (Figure 3) and matK gene (Figure 4). The trees indicated that there were two major branches of group of selected pine species together. This study evaluated usefulness of rbcL and matK genes for molecular identification and phylogenetic study among various species of Pinus. Cluster analysis of the data obtained from rbcL gene belonging to ten Pinus species revealed four clusters. First Cluster included P. Wallichiana (PW), P.Elliottii (PEl) and P.Greggii (PG). Second cluster included P. Thunbergii(PT),P. echinata (PE) and P.Merkusii (PM). Third cluster included P.taeda (Ptd) and P. Khasya (PK) while fourth cluster included P. Roxburghii(PR) and P. Patula (PP) as shown in Figure 3. Similarly cluster analysis of the data obtained from matK gene belonging to ten Pinus species revealed four cluster and one outgroup. First Cluster included P. Thunbergii(PT), P.Greggii (PG) and P.Elliottii (PEl) Second cluster included P. Roxburghii(PR) and P.Merkusii (PM), third cluster included P. Wallichiana (PW) and P.taeda (Ptd) while fourth cluster included P. Patula (PP) and P. Khasya (PK). P.echinata remained out clustered in this analysis (Figure 4).
Discussion
Analyzing the amounts of nucleotide and patterns of nucleotide variation between and within species is important to understand the mechanisms of evolution by which processes of genetic polymorphisms within species become transformed into genetic divergence between species and genetic diversity is maintained. Such diversities are influenced by evolutionary processes, such as selection, recombination, mutation and population structures.
In one of the earliest studies by Govindaraju, et al. 1992 [9], phylogenetic study amongst 30 species of the Pinus, P. taeda, P. echinata, and P. elliottiiwere classified under the section Pinea and sub section Australes. The study also revealed that majority of the studies had maximum likelihood relationship with the species of Pinus . Majority of the species of Pinus fall under the lineage of Ponderosae, Oocarpae, Contortae, Australes, and Sabinianae. Wagner, et al.[10] also conducted phylogenetic study among Pinus sp., Pinus echinata, Pinus elliottii, Pinus palustris, and Pinus taeda. Phylogenetic studies on the basis of the study by Little and Critchfield [4] found that these species of Pinus had distinctive relations among them and the clades or sub sections mainly Australes and Ponderosae have common lineage. On the other hand, P. wallichiana was reported under the sub section StrobusP. gerardiana under the sub section Gerardianae; P. Khasya and P. Thunbergiiunder Pinus, P. merkusii under subsection Pinaster, P. Roxburghiiunder Pineae, P. echinata under Australes, P. patula under Oocarpae according to a phylogenetic study by [5]. The study found a distant relationship among the sub sections Strobi, Pinus, Pineae, Pinaster, Oocarpae, and Australes. However, Oocarpae, and Australes indicated the closest lineage amongst the Pinus sp. Furthermore, the phylogenetic study found that Pinus, Pineae and Pinaster subsetions were closely related to each other but distant relationship to Strobi for P. wallichiana and Oocarpae, and Australes for P. echinata and P. patula. However, these species were found to spread across North and Central American subsections to the Himalayas with common lineage that brings them under the same phylogeny.
Krupkin, et al. [11] also conducted phylogenetic analysis among 18 species of Pinus and used Wagner parsimony analysis for exploring the phylogeny of the Pinus sp. The study found that the clades Contortae, Ponderosae, Sabinianae, Australes, and Oocarpae consisted of majority of the Pinus sp. such as P. taeda, P. echinata, P. gregii, P. patula, P. elliotti and others. The species of these sub sections were in close parsimony with respect to anatomical structures and genetic structures. The study also found that the species under the sub section Contortae, shared distinctive relations with P. taeda or the other Pinus sp. of Ponderosae and Australes. Furthermore, the study argues the phylogenetic distribution of few Pinus species by [4] like Leiophyllae clade was found to have close lineage with Ponderosae, Oocarpae, Contortae, Australes, and Sabinianae.
Lately, [12] and [13] used matK, rbcL, and other genes to understand the phylogeny of the Pinus sp. According to [12] P. taeda, P. echinata, P. greggii, P. patula, and P. elliottiiwere all classified into the sub section Australes of the Pinus. P. greggii was the only species that had minimal genetic linkage to P. taeda, P. echinata, P. patula, and P, elliottii, whereas P. taeda and P. patula were closely related and P. echinata and P.elliottii showed close relation as they all belonged to the North and Central American regions. However, P. Thunbergiiwas classified under the sub section Contortae and had low genetic linkage to P.taeda, P.echinata, P.greggii, P. patula and P.elliottii as they were found in the Himalayan region. Similarly, P. greggii and P.patula were positioned under Australes subsection with high genetic lineage, whereas P.thunbergii was positioned under Contortae.
Singh and Thapliyal [14] analyzed seed sources for genetic variation amongst exotic and indigenous Pinus sp. and the result presented a significant diversity in height, length, the width of cone and seed. The overall study result presented the heritability values of seed weight associated with maximum genetic gain. Such genetic traits disclosed a strong genetic control that revealed phenotypic and genotypic variations among the seed and cone are a result of genetic diversity [14]. The genetic variation occurred due to genotypic and environmental interaction. This concluded that the Indian species P. roxburghii, P. wallichiana, P. kesiya, P. gerardiana, P.merkusii, P. thunbergii, and P. clausa are closely related to each other with respect to needle anatomy and morphology as well as on the basis of genetic structure. In this study, out of the 10 taxa examined nine belong to subgenus Pinus and one to subgenus StrobusWith respect to anatomical structure of the leaf, two subgenera were easily distinguishable by the number of vascular bundles in their needle. Although various morpho-anatomical classification schemes have been proposed for the sections and subsections of subgenus Pinus as well as subgenus Strobusthe relationships among the subsections and their evolutionary processes are still being debated [3,14-18]. Due to large number of morpho-anatomical characters and presence of high levels of homoplasy in many of these morpho-anatomical characters, several differences among classification schemes have been encountered. Thus, a comprehensive treatment of phylogenetic analysis of both the subgenus is hampered by the inadequacy of discrete characters, which are also scarce in the genus Pinus compared to other plant groups [15]. Several molecular studies have been conducted on the species of both subgenus Pinus and subgenus Strobus and these studies revealed a large genetic dissimilarity between the two subgenera and within various species of these subgenera. as well as little genetic variation in subgenus Pinus [7,9,11,19]. However, these studies have been limited in terms of taxonomic sampling and/or geographic scope, particularly in subgenus Pinus and pine species growing in Europe and American continents. The study by [11] using chloroplast DNA (cpDNA) analyzed 18 Diploxylon pines species of North America. Other studies such as analysis of ITS sequences by [5] involved a broad sampling of Pinus subsections and covered a wide range of geographic regions. Emphasis on native species of Eurasian pines and their phylogenic relationship with other species growing in their vicinity after their introduction from other continents due to their economic interest is missing. However ITS data generated in the study strongly supported the hypothesis of the presence of a distinctive group of North American pines.
Conclusion
Further studies involving as many as species possible using multiple marker, including both subgenera and all their sections as well as subsection are need to be studied to develop a complete database that can be used for a full proof classification and identification of Pinus up to species level.
Authors contribution
LS was a major contributor in writing the manuscript, GS and PCV proposed the research idea, conceptualized the research design and prepared the final manuscript including the original experimental work, and data recording. PJ, RS, SP and AS was involved with material collection and identification.
The authors thank Head, Department of Botany, University of Lucknow for providing necessary research facilities.
- Farjon A (1984) Pines: drawings and descriptions of the genus Pinus. EJ Brill and W Backhuys, Leiden, Netherlands. Link: https://bit.ly/3zUL4JV
- Price RA (1982) The genera of Pinaceae in the southeastern United States. J Arnold Arbor 70: 247-305.
- Mirov NT (1967) The Genus Pinus. The Ronald Press Company. New York, USA. 602. Link: https://bit.ly/3gNLB8W
- Little EL, Critchfield WB (1969) Subdivision of the genus Pinus (Pines). USDA Forest Service Miscellaneous Publication 1144, Washington, DC. Link: https://bit.ly/3wNymKU
- Liston A, Robinson WA, Piñero D, Alvarez-Buylla ER (1999) Phylogenetics of Pinus (Pinaceae) based on nuclear ribosomal DNA internal transcribed spacer region sequences. Mol Phylogenet Evol 11: 95-109. Link: https://bit.ly/3cXARlJ
- Wang XQ, Tank DC, Sang T (2000) Phylogeny and divergence times in Pinaceae: evidence from three genomes. Mol Biol Evol 17: 773-781. Link: https://bit.ly/3vQUClI
- Wang XR, Tsumura Y, Nagasaka H, Szmidt HE (1999) Phylogenetic relationships of Eurasian pines (Pinus, Pinaceae) based on chloroplast rbcL, matK, rpl20-rps18 spacer and trnV intron sequences. Am J Bot 86: 1742-1753. Link: https://bit.ly/2SSrKfw
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876-4886. Link: https://bit.ly/2TRsSAj
- Govindaraju D, Lewis P, Cullis C (1992) Phylogenetic analysis of pines using ribosomal DNA restriction fragment length polymorphisms. Plant Syst Evol 179: 141-153. Link: https://bit.ly/2TO0yyC
- Wagner DB, Nance WL, Nelson CD, Li T, Patel RN, et al. (1992) Taxonomic patterns and inheritance of chloroplast DNA variation in a survey of Pinus echinata, Pinus elliottii, Pinus palustris, and Pinus taeda. Canadian Journal of Forest Research 22: 683-689. Link: https://bit.ly/3j2e0JB
- Krupkin AB, Liston A, Strauss SH (1996) Phylogenetic analysis of the hard pines (Pinus subgenus Pinus, Pinaceae) from chloroplast DNA restriction site analysis. American Journal of Botany 83: 489-498. Link: https://bit.ly/3zOsNO3
- Hernández-León S, Gernandt DS, de la Rosa, Jardón-Barbolla L (2013) Phylogenetic relationships and species delimitation in Pinus section Trifoliae inferrred from plastid DNA. PloS One 8: e70501. Link: https://bit.ly/3j1vTsm
- Gernandt DS, López GG, García SO, Liston A (2005) Phylogeny and classification of Pinus. Taxon 54: 29-42. Link: https://bit.ly/3cZNBIT
- Singh O, Thapliyal M (2012) Variation in cone and seed characters in blue pine (Pinus wallichiana) across natural distribution in western Himalayas. Journal of Forestry Research 23: 235–239. Link: https://bit.ly/3gLqCDF
- Farjon A, Styles BT (1997) Pinus. Flora Neotropica Monograph 70.New York Botanical Garden, NY.
- Rushforth K. Conifers., 1987. London.
- Klaus W (1989) The Mediterranean pines and their history. Plant Syst Evol 162: 133-163. Link: https://bit.ly/3qooDIE
- Perry JP (1991) The pines of Mexico and Central America. Timber, Portland, Oreg 231. Link: https://bit.ly/35HJz3H
- Moran GF, Smith D, Bell JC, Appels L (1992) The 5S RNA genes in Pinus radiata and the spacer region as a probe for relationships between fPinus species. Plant Syst Evol 183: 209–221. Link: https://bit.ly/2STR9oX
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
