Open Journal of Proteomics and Genomics
Pillaging plucking plundering ransacking proteomes via CPLL technology
Pier Giorgio Righetti1* and Egisto Boschetti2
2Scientific Consultant, JAM Conseil, 92200 Neuilly-sur-Seine, France
Cite this as
Righetti PG, Boschetti E (2023) Pillaging plucking plundering ransacking proteomes via CPLL technology. Open J Proteom Genom 8(1): 001-010. DOI: 10.17352/ojpg.000012Copyright License
© 2023 Righetti PG, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.No proteome can be considered “democratic”, but rather “oligarchic” since a few proteins dominate the landscape and often obliterate the signal of the rare ones. That is the reason why most scientists lament that, in proteome analysis, the same set of abundant proteins is repeatedly seen. Current pre-fractionation techniques, one way or another, are besieged by problems, in that they are based on a “depletion principle”, i.e. elimination of unwanted species. Yet “democracy” calls for giving “equal rights” to everyone. One way to achieve that would be the use of libraries of combinatorial ligands coupled to spherical beads. When these beads are contacted with complex proteomes (e.g., human urines and sera, egg white, any cell or tissue lysate) of widely differing protein composition and relative abundances, they are able to “normalize” the protein population, by sharply reducing the concentration of the most abundant components while simultaneously enhancing the level of the most dilute components. It is felt that this method could offer a strong step forward in bringing the “unseen proteome” (due to either low abundance and/or presence of interferences) within the detection capabilities of current proteomics detection methods. Examples are given of the normalization of human urine and sera samples, resulting in the discovery of a host of proteins previously unreported. These beads can also be used to remove host cell proteins from purified recombinant proteins or proteins purified from natural sources that are intended for human consumption. These proteins typically reach purities of the order of 98%: higher purities often become prohibitively expensive. Yet, if incubated with Combinatorial Peptide Ligand Libraries (CPLL), even these impurities can be effectively removed with minute losses of the main, valuable product.
Abbreviations
TUC: Thiourea, Urea, Surfactants; CB: Coomassie Blue; LDS: Lithium Dodecyl Sulfate; EB: Equalizer Beads
Introduction
Tiselius, with his monumental moving boundary apparatus [1], as well as cellulose Acetate Electrophoresis (CAE) [2], in the analysis of human sera for early diagnosis of different diseases, could detect barely 6-7 major components in sera. When biochemists started analyzing sera via 2-D mapping, the picture changed drastically: at least 600 spots could be revealed scattered in the pI/Mr plane, via silver staining [3]. Yet, even with the advent of 2-D mapping, no new markers of diseases could be found. The reasons are outlined in a paper by Pieper, et al. [4]: it is nearly impossible to detect any early potential marker of disease as long as the whole serum is analyzed since the most abundant proteins will obliterate the signal of the rare ones (the dynamic range of plasma protein concentrations comprises some ten orders of magnitude or more!). Even when removing the nine most abundant human serum proteins (albumin, IgG, haptoglobin, transferrin, transthyretin, α1-antitrypsin, α1-acid glycoprotein, hemopexin, and α2-macroglobulin the end results were not so exciting: no new markers of disease could be found. However, via this process, these authors could finally visualize several proteins present in sera in < 10 ng/mL concentrations, such as interleukin 6, cathepsin and peptide hormones [4].
Pre-fractionation would be the key to success: a host of pre-fractionation techniques have been reported over the years, as reviewed in [5-9]. The approach that is gaining momentum, especially in the analysis of biological fluids, such as plasma, sera, cerebrospinal fluid, and urines, is sequential or simultaneous immuno-affinity depletion of the most abundant proteins present therein, as outlined in [4]. Even this approach might not be enough for gaining access to the “deep proteome”. Although depletion of these 9 abundant proteins represents the removal of 90% of the overall protein concentration, the vast number of serum proteins in the remaining 10% protein concentration remains to dilute and the improvement of detection of rare proteins is quite disappointing, as evidenced by Echan, et al. [10], who suggested the use of antibody columns that could deplete at least 18-22 of the most abundant proteins, which comprise 98-99% of the total serum proteins. In fact, Huang, et al. [11] reported the use of IgY microbeads able to capture and efficiently remove the top set of 12 most abundant plasma proteins, whereas depletion methods based on chemical agents such as Cibacron Blue perform poorly: according to Zolotarjova, et al. [12] this chemical removed a major portion of the targeted albumin protein but also removed many other low-abundance proteins from serum, as reported in [13].
Yet, even the more specific immuno-depletion approach might induce parasitic co-depletion of a number of proteins that might be physiologically bound to the proteins captured by immuno-subtraction. A case in point is albumin depletion: according to Zhou, et al. [14], when albumin is immuno-depleted, another 63 proteins, bound to albumin, are co-depleted as well. Another quite disappointing result: after immuno-depletion, although the number of spots displayed in a 2D gel is substantially increased, the number of new proteins identified is quite small, due to the emergence of newly visualized spots representing isoforms of the now most abundant proteins. And, even after any immuno-subtraction technique, the rare and very-rare proteins are still invisible, since, by this process, they are not concentrated and thus remain below the detection limit of most analytical methods.
The CPLL approach here reviewed might turn out to be an important step for bringing to the limelight the “hidden proteome” and discovering several new biomarkers for clinical chemistry analysis. A basic article, outlining the synthesis of the beads and some of their fundamental properties [15], as well as reviews describing the very basic concepts [16,17], have appeared. Here, we offer an in-depth review of the physico-chemical properties of this ligand library, together with some applications that demonstrate the unique potential of this approach.
Chemistry and synthesis of the peptide ligand library
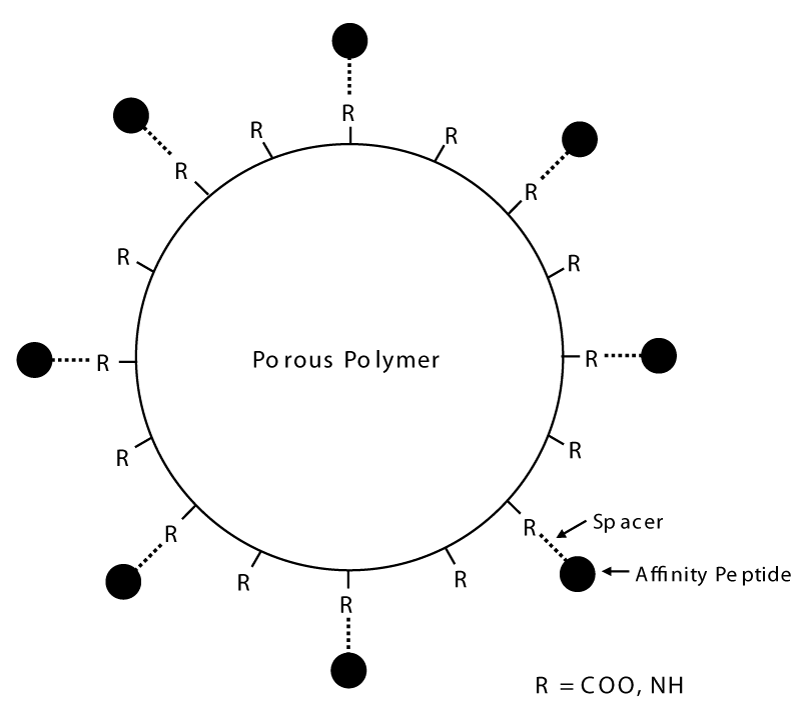
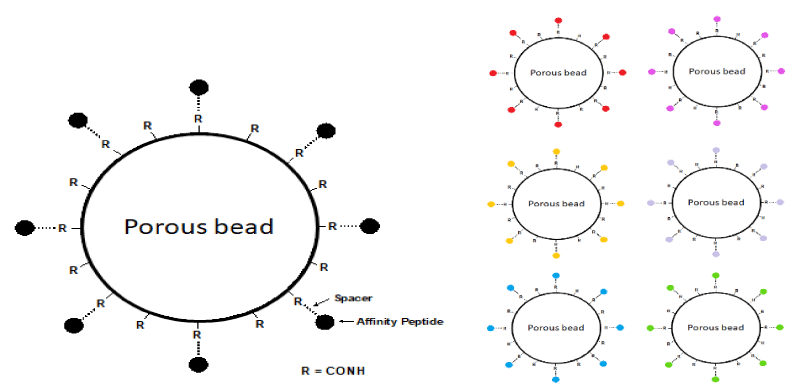
The solid phase combinatorial libraries of hexapeptides are synthesized via a short spacer on poly (hydroxy methacrylate) beads, via a modified Merrifield approach [18-21]. Figure 1 gives a pictorial representation of the structure of the beads. The ligand’s density in the bead porous structure can reach a concentration of 50 pmoles per mL. This amounts to a ligand density of ca. 40-60 µmoles per mL of bead volume. Every single bead, thus, has millions of copies of a single, unique ligand and each bead, potentially, has a different ligand from every other bead. Considering that, for the synthesis, the 20 natural amino acids are used, this means that the library contains a population of linear hexapeptides amounting to 206, i.e. 64 million different ligands. Such a vastly heterogeneous population of baits means that, in principle, an appropriate volume of beads could contain a partner able to interact with just about any protein present in a complex proteome. Using a hexapeptide ligand for establishing an affinity interaction might be considered to represent a rather weak binding event; however, experience has demonstrated that indeed such a complex can be of very high affinity that would require very strong elution conditions.
Types of bonds engendered between captured proteins and combinatorial ligands
The types of bonds that are established between a protein and its partner in the hexameric population are the classical ones that stabilize the conformation of macromolecules, i.e. weak interactions that are at least one order of magnitude less strong than that of covalent bonds. Such interactions, as summarized in Figure 2A [22], describe how atoms or groups of atoms are attracted or repelled to minimize the energy of conformation. They can be grouped into ion-ion, hydrogen bonding, dipole-dipole, dispersion, hydrophobic interactions as well as van der Waals interactions. These are, in general, distance-dependent interactions, with the energies being inversely proportional to the distance r or to some power of the distance (r2, r3, etc.) separating the two groups. As the power of the inverse distance dependency increases, the interaction approaches zero more rapidly as r increases and thus becomes a short-range interaction. Figure 2B gives the energy of these interactions as a function of the distance r between the two interacting atoms. It can be appreciated that ion-ion interaction, where n = 1, is a long-range interaction, still appreciable at a distance of at least 1 nm. Conversely, London dispersion forces (n = 6) and steric repulsion forces (n = 12) are classified as very short-range interactions, since they approach E = 0 much more rapidly, typically within 0.2 nm to 0.25 nm.
The expression for the energy of long-range interactions are all inversely related to the dielectric constant of the medium and are thus weakened in a highly polarizable medium such as water. The composition of the medium will also affect other important weak interactions, such as hydrogen bonds and hydrophobic interactions. This is why, when capturing proteins with the hexameric ligand library, the process is conducted under native conditions, i.e. at physiological pH and at an ionic strength compatible with such native conformations. Denaturing media (such as TUC, thiourea, urea, and surfactants, commonly adopted in 2D map analysis) [23,24] will not be amenable to treatment with CPLLs, since TUC is one of the typical eluants of species captured by these beads.
Another important factor in the linkage between proteins and the hexameric peptide baits is hydrophobic interaction. The most hydrophobic Amino Acids (AA) are, in the order, of Ile, Val, and Leu. Paradoxically, Trp and Tyr, as measured by the hydropathy index, as obtained by water/octanol partitioning, are even more hydrophilic than Gly, Thr and Ser, one due to its phenolic –OH (Tyr) and the other due to its hetero-aromatic ring (Trp) [25].
Elution paradigms
Having singled out the main mechanism of interaction between native proteins and CPLLs, it is now possible to devise elution procedures enabling the recovery of adsorbed proteins. Elution could be implemented either as a single process (with a strong eluant able to discharge all adsorbed material) or as a cascade process (with eluants of increasing strengths). The latter protocol would have the advantage of permitting a further sub-fractionation of the “normalized” proteome without using sequences of other chromatographic methods. For ion-ion dominating interactions, a classical eluant would be 1 M NaCl, as customarily done in ion exchange chromatography. This process, in general, should allow the recovery of proteins in a native form, retaining biological activity.
For breaking mildly hydrophobic interactions, one could resort to 50% ethylene glycol [26]. However, this eluant is quite mild and the population of proteins released is quite restricted; thus it has not been routinely adopted in our protocols. As described for serum proteins [27], hydro-organic solutions also contribute to eluting hydrophobic proteins. Another type of elution could be via 200 mM glycine-HCl, at pH 2.5: this eluant is typically adopted to disrupt tenacious interactions possibly related to conformational structures, such as those occurring between antigens and antibodies in an immuno-affinity column [28]. Very low pHs contribute to significantly deform protein epitopes reducing thus the ability to interact with hexapeptide ligands.
TUC (2 M thiourea, 7 M urea, 4% CHAPS) appears to be an excellent eluant for proteins adsorbed onto CPLL beads. It is a mixed-mode eluant, able to disrupt simultaneously hydrogen bonds as well as hydrophobic associations; it releases a large population of adsorbed material. Concentrated urea solutions at acidic or alkaline pHs could also be used with an almost quantitative desorption efficacy. For eluting proteins en masse, one could use 6 M guanidine HCl (GuHCl), pH 6. Due to its strong chaotropic effect and its high ionic strength this solution is considered a general eluant, able to disrupt all bonds and reduce all proteins to random polymer coils [29]. GuHCl can be used as the sole elution step if all proteins have to be desorbed at once, or as the final step, for eluting the proteins most tenaciously binding to the solid phase.
The physico-chemical mechanism of protein capture by CPLL is frequently based on complex interactions, it is not always easy to find a universal elution method. Thus a paradigm change is represented by the direct on-bead trypsinization of captured species. The availability of whole proteins is in fact not necessary when the game is to identify the proteome components. Such an approach produces mass spectrometry that can easily be interpreted. Most of the time the trypsinization of one protein ends up in many peptides whereas only two peptides are sufficient to identify the protein origin. This methodology called MudPIT (Multidimensional Protein Identification Technology) was first described at the beginning of this century and appears very convenient as it saves time compared to protein elution followed by electrophoresis fractionations and protein identification. Results using CPLL technology have been gradually reported with very interesting results around the discovery of proteins of very low abundance. In 2011 Meng, et al. [30] and Fonslow, et al. [31] described this method to detect biomarkers of breast cancer from human plasma on the one hand and improve proteomic metrics from HeLa cellular lysates, on the other hand. Improved reproducibility has also been reported. Other authors adopted this protocol for the analysis of several biological materials such as a synovial fluid with an improvement of the number of proteins identified of more than 30% compared to the current methodology [32]. The bead trypsinization prior to mass spectrometry was further optimized and improved by extending the digestion time and also by making a pre-digestion with Lys-C endopeptidase [33]. It is here important to say that even if some peptides are still strongly attached to the beads after digestion, they do not degrade the mass spectrometry analysis because the number of peptides needed for protein identification is limited to two. The increased number of detected proteins, the lower risk of protein losses due to too hard elution conditions, and the simplified protocol with better reproducibility should play in favor of the on-bead protein digestion option, especially when looking for protein markers discovery and for the detection of protein impurities present in biopharmaceuticals.
The CPLL technology versus depletion procedures
It is well-known that albumin, and a few other very high-abundance proteins in plasma, represent a challenge for the proper detection of other low-concentration proteins. Concurrently, albumin is a source of trouble in mass spectrometry since it induces signal suppression of a number of other species, which by consequence are not detected. In this situation, a quite common approach is to remove high-abundance species prior to analysis.
Depletion is nothing more than a separation of proteins by solid phase adsorption and this fact necessitates some focused considerations that are related to well-established rules in terms of thermodynamics, kinetics, and binding capacity. Depletion of one abundant protein or its adsorption on chromatographic support is directly dependent on the binding capacity for the protein in question. Cibacron Blue, commonly used for the depletion of albumin, has a binding capacity between 10 mg/mL and 30 mg/mL of resin. Proteins collected in the flow-through are generally diluted by a factor of 2 - 2.5. Considerations around dilution are even worst with lower binding capacity resins. These latter generally have a binding capacity of a few mg/mL for a single protein (between 1/20 and 1/5 of Cibacron Blue for instance for the depletion of albumin). This means that the volume of the sample compared to the volume of the sorbent is very small.
The situation should be improved with the use of biologically specific ligands; this is the case of protein A and Protein G for the elimination of IgG antibodies. Even here the operation results in the elimination of additional proteins. A better example is represented by highly specific depletion with antibodies. However, the binding capacity is very limited and its use is restricted to plasma proteins only. The cost of immunosorbents is also very high compared to current depletion methods.
Figure 3 illustrates an SDS-PAGE analysis: when the depletion is operated with synthetic ligands such as Cibacron Blue, the elimination of albumin is not complete and other species are not visible any longer. When using an immunodepletion using six immobilized antibodies, the remaining protein sample is quite clean but the number of detectable proteins has not really changed. Conversely, the CPLL technology shows the largest number of protein bands over every other depletion method.
Analysis of human body fluids with CPLL
Since a number of applications of CPLL beads to different biological samples have been already described [15-17], we will concentrate here on other applications, particularly in regard to human body fluids. Clinical chemist research has been focused on finding, especially in body fluids – plasma, urine, tears, lymph, seminal plasma, milk, saliva, spinal fluid – new indicators or markers for disease. Even body fluids are not immune from severe problems that have hampered the discovery of novel markers; e.g., both plasma and serum exhibit very high variations in individual protein abundances, typically of the order of 1010 or more, with the result that, in any typical Two-Dimensional (2D) map, only the high-abundance proteins are displayed [4]. In the case of urines, the problems are further aggravated by their very low protein content requiring a concentration step of 100 to 1000 fold, coupled with their high salt levels, demanding their concomitant removal prior to analyses [34].
Analysis of human urine proteins
Considerable research efforts have been devoted to mapping the human urinary proteome since this is perhaps the only one that can be collected in a fully non-invasive manner and in large volumes repeatedly and for extended periods of time [35-45]. Although the vast majority of them have exploited 2D maps, a few reports have also described 1D and 2D chromatographic approaches [35,44,45]. 2D map analysis has been already exploited in bladder cancer [46,47], Bence Jones proteinuria [48,49], rheumatoid arthritis [50], urinary tract infections and glomerular or non-glomerular diseases [51,52], chronic exposure to cadmium [53], characterization of urinary apolipoproteins and monitoring adaptive changes in unilateral nephrectomy [54] and searching for novel candidate markers for prostatic cancer [55]. Most of these approaches require a large number of steps for urine preparation prior to 2D mappings, such as precipitation with protamine sulfate, removal of glycosaminoglycans, several dialysis steps, lyophilization, gel filtration, even immuno-subtraction of the most abundant proteins and other pre-fractionation tools [16,36,56].
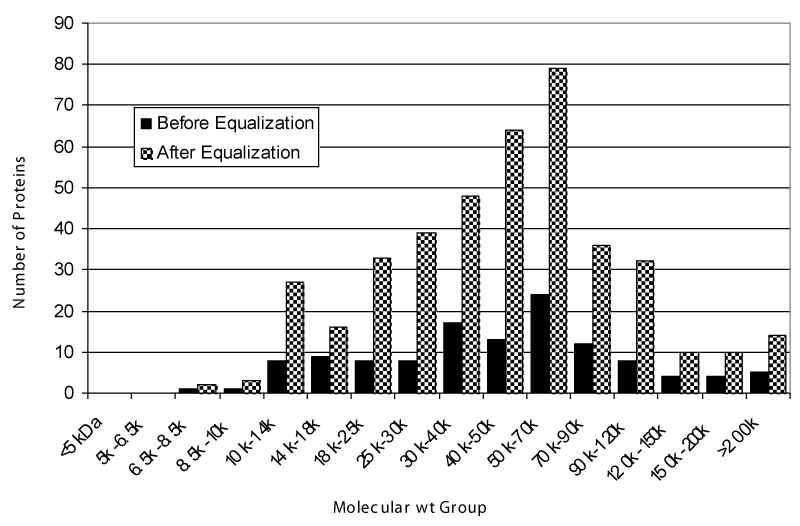
CPLLs were thus adopted in urine analysis [27]. A total of 1.6 L of urines was collected from eight healthy young donors, processed, and reduced to a volume of 22 mL dissolved in 25 mM phosphate buffer, pH 7.0. This final volume was adsorbed onto 1 mL of beads, which were then eluted first with TUC (2.2 M thiourea, 7.7 M urea, 4.4% CHAPS) and then with 9 M urea at pH 3.8. The results of the 2D mapping, as compared to untreated controls, are displayed in Figure 4. The dynamic range reduction effect, with a concomitant massive increment of polypeptide spots over the entire gel surface, is evident in the 2D maps of the treated samples. The first eluate (TUC) exhibits many more spots in the entire pH interval as compared with control urine. The second eluate, although displaying a significantly lower number of spots, shows only a limited redundancy with the TUC eluate, most desorbed proteins being specific to the second elution step. These three samples were then subjected to FT-ICR (Fourier transform, ion-cyclotron resonance) mass spectrometry analysis. Control urines revealed a total of 96 unique gene products. The TUC eluate allowed the identification of 334 unique protein species and the second eluate of an additional 148 species. This gives a total count of 471 unique protein species identified in urines. The bar graph of Figure 5 gives the increment in species obtained in the sum of the two eluates, as compared with the control, while simultaneously expressing their Mr distribution. These results are impressive when compared to the best data available in the literature, by using much more complex technologies and experimental protocols. Summing up: (i) Pieper, et al. [35] reported 150 unique protein annotations; (ii) Spahr, et al. [43] described 124 gene products; (iii) Oh, et al. [36] listed 113 different proteins; (iv) Pang, et al. [34] found 103 unique species; (v) Castagna, et al. [27] detected 471 unique gene products via the CPLL technology.
Analysis of human serum proteins
The serum is still one of the biological fluids of utmost interest from a clinical-chemistry point of view [57,58]. Up to 2004, the only extensive data set available on serum proteins was the one of Anderson, et al. [58], with a compilation of 1175 non-redundant species. It was quickly superseded by the HUPO Plasma Protein Project (PPP). The PPP was started in the year 2002 as a network of 35 collaborating laboratories. A core set of 3020 serum plasma proteins could be generated and is now available on public databases (www.bioinformatics.med.umich.edu/hupo/ppp) [59]. This dataset is composed of unique gene products identified with two or more peptides, thus predicted to be correct with a confidence of the order of 0.90 to 0.95. The data set contains an additional 6484 unique proteins identified via only a single peptide, bringing the total to 9504.
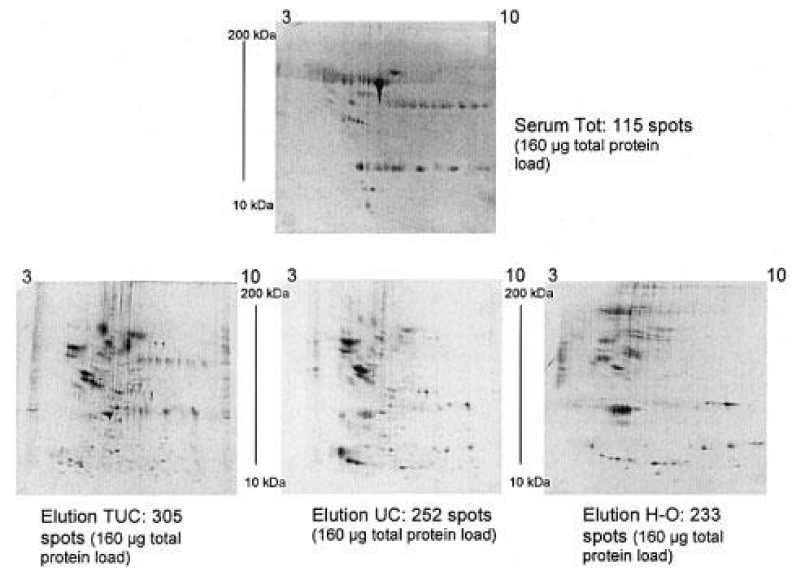
What portion of serum proteome could be detected within a single, simple experimental protocol exploiting CPLLs? 300 mL of serum were processed and subjected to adsorption with our ligand library. Upon elution with 3 different eluants, the 2D maps of Figure 6 could be obtained, showing a large increment of spots in both eluates, as compared to control sera. Upon elution and analysis via FT-ICR mass spectrometry, a total of 4802 unique gene products could be recognized [60]. Our data compare favorably with the HUPO list of plasma proteins, totaling 9504 species. Additionally, > 3000 species were unique to our list, and are believed to represent the “deep proteome” so far undetected.
Purification of r-DNA products
Recombinant DNA (r-DNA) products are becoming one of the most important families of therapeutic agents, due to the possibility of production on a large scale in host organisms [61]. One of the major dilemmas associated with the production of r-DNA proteins for human consumption is their extraction and purification from very crude feedstocks. In principle, such recombinant proteins should be purified to homogeneity, so as to avoid undesired side effects when they are co-injected with traces of protein impurities that can be immunogenic. It is very laborious to achieve purity levels better than 99% [62,63]. The last purification step is prohibitively expensive and leads to severe losses of a valuable biopharmaceutical product. This is the reason why a protein purification process, designed for biopharmaceuticals starting from crude feedstocks, is relatively complicated. The first chromatographic separation step of the process is based on the selectivity of the resin for the target protein. In that way, it is possible to concentrate the protein of interest and get rid of the maximum amount of protein impurities. This step is generally named “capture” [64]. If the capturing resin is based on a high-affinity ligand the resulting purification factor in a single step could be very substantial. This is the case for the capture of monoclonal antibodies when using a Protein A column [65] where the purity of the target protein can go up to 95% - 98%. The pre-purified protein from the capture phase is then submitted to an intermediate separation process capable to produce fractions where the protein of interest is collected in one or two of them [66-68]. In order to reach purity levels capable to meet regulatory requirements, however, it is necessary to add at least another step called “polishing” capable to eliminate all remaining impurity traces.
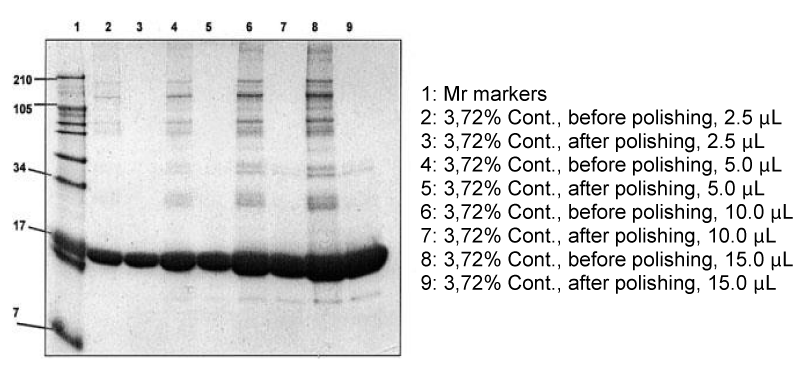
Products in the market rarely achieve a purity degree better than 98% - 99%. Pharmaceutical companies, in general, are able to identify one or a few major contaminants and this is the pedigree of the products available on the market for human consumption. The situation, though, could be much worse than that. We have set up an experimental model for monitoring what could be achieved when proteins, contaminated with different amounts of impurities, would then subjected to a cleaning step with our ligand library. The protocol comprised a 10 mg/mL solution of very pure myoglobin, re-purified on a column of Q HyperD in 25 mM phosphate buffer, pH 7. This solution, containing 400 mg of myoglobin, was contaminated with 10 μL of serum protein eluate from CPLL beads the removal of contaminants was monitored by SDS-PAGE. The results are shown in Figure 7. After this cleaning step, the vast majority of contaminants are removed. A complete polishing could be obtained by re-subjecting the final products to a second CPLL treatment [69]. This process has a double valency: on the one hand, it allows effective removal of all impurities; on the other hand, it permits concentration and purification of these impurities so as to allow proper characterization of all of them.
“Missing in action”
We consider here some important experimental variables.
Concentration limits: The first question is about the lowest concentration limit of proteins in body fluids and cell lysates that can be detected. With a proviso, though, that this limit might not exist, since it will strongly depend on what is the total sample volume available for treatment. Given enough sample volume, even those ultra-rare proteins present in a few copies per cell could be rendered visible. The association/dissociation constants of proteins to the library of ligands will, ultimately, be the driving force behind the capturing ability of the beads. Those will depend on a number of variables, such as temperature, ionic strength, and pH value of the buffering media.
Protein losses: To which extent the components of a given proteome will be lost, either due to the lack of appropriate ligands or to the too-weak Ka values of peptide ligands towards a number of components has to be considered. Data comparing proteins identified in control versus CPLL samples suggest that some 3% to 7% of the total protein population might be lost.
Spurious binding: Is the spurious binding of the proteins to other components of the beads (e.g. matrix polymer qualified as non-specific binding) than with their complementary bait ligand counterpart taking place? Experiments with the beads lacking the hexameric peptide baits suggest that parasitic binding to the organic polymer used as substratum is virtually nihil.
Unbalanced binding: Whether abnormal binding of proteins to the bead library, leading to non-normalized situations occurs or not, is another non-completely answered the question. It is unrealistic to think that all proteins will be well-behaved toward the adsorbing ligand library. Working with sera, we have found for instance that apolipoprotein A1 (Apo A1) is greatly enriched as compared to all other serum components, rendering it the most abundant component after the treatment. We have no explanation for that, except to note that Apo A1 possesses a large number of binding sites for several components. Thus, it is quite possible that it recognizes more that one hexapeptide ligand, saturating an abnormal amount of sites in a larger bead population as compared to other, well-behaved proteins.
Length of the bait: Another parameter that has an effect on the reduction of the dynamic concentration range of proteins is the length of the bait peptide. Although current CPLLs carry a population of hexameric ligands, lengthening it to a heptamer, while greatly expanding the diversity of the baits (from 64 million to 128 billion), thus reducing the chances of losing parts of the proteome components, will surely strengthen the value of the association constant.
Bead size: Finally, the bead size would play a role in the capturing process, but data are not well-known yet. Clearly, experimenting with smaller and smaller beads will allow the processing of lower volumes of precious biological fluids, since the bait density per unit of beads volume will increase. However, performing a classical combinatorial synthesis of ligands onto very small beads (e.g., 5 µm and below) greatly changes the thermodynamics and kinetics of the interaction along with binding capacity and may additionally complicate the synthetic process.
Hunting for biomarkers: Would this technology be amenable to biomarker hunting, given its propensity for equalizing concentration differences? In principle this should be possible, as long as the biomarker sought, in the control vs. the pathological sample, is in such minute amounts as not to saturate the beads. Under these conditions, the relative concentration differences should remain rather unchanged and thus the diagnostic value should still be there. In this context, hundreds of publications focusing on biomarker discovery using CPLL are available describing single protein marker discoveries as well as entire panels of proteins witnessing their utmost interest in diagnostics and therapy.
Conclusion
We hope we have given here a fair survey of the capability of the CPLL technology in dealing with the vastly diverging protein concentrations in any proteome isolated from living organisms. We have offered a brief survey of data from two human fluids, urines and sera, that have important implications for the discovery of biomarkers of most pathologies. We hope the readers are convinced by the great potential of this technique that has allowed, for the first time, and with simple manipulations, to truly uncover the hidden proteome.
Integration of this technology with classical or novel fractionation methods will increase the capability to discover novel proteins of diagnostic interest. In addition to its implications in proteome analysis, the described approach will also play a very important role in the pharmaceutical and biotech industry. For the former industry, this technology will contribute to finding low protein expression as a response to newer therapies with possibly important implications for drug indications and reactions. In the case of the Biotech industry, the technology will contribute helping to remove from r-DNA products, meant for human consumption, the last 2% - 3% of impurities that are still contaminating all products presently in the market, impurities that might have severe adverse effects for patients using such products not just for single treatment but as a lifetime cure.
We thank Drs. J. Rappsilber, L. Sennels, D. Cecconi, and A. Castagna for their valuable help in urine and sera analyses and Drs. L. Guerrier and F. Fortis for providing data on protein depletion methods.
- Tiselius A. New Apparatus for Electrophoretic Analysis of Colloidal Mixtures. Transactions of the Faraday Society. Trans. Faraday Soc. 1937; 33: 524-531.
- Kohn J. Clin Chim Acta. 1957; 2:297-305.
- Hochstrasser D, Augsburger V, Funk M, Appel R, Pellegrini C, Muller AF. Immobilized pH gradients in capillary tubes and two-dimensional gel electrophoresis. Electrophoresis. 1986; 7: 505-511.
- Pieper R, Gatlin CL, Makusky AJ, Russo PS, Schatz CR, Miller SS, Su Q, McGrath AM, Estock MA, Parmar PP, Zhao M, Huang ST, Zhou J, Wang F, Esquer-Blasco R, Anderson NL, Taylor J, Steiner S. The human serum proteome: display of nearly 3700 chromatographically separated protein spots on two-dimensional electrophoresis gels and identification of 325 distinct proteins. Proteomics. 2003 Jul;3(7):1345-64. doi: 10.1002/pmic.200300449. PMID: 12872236.
- Righetti PG, Castagna A, Herbert B. Prefractionation techniques in proteome analysis. Anal Chem. 2001 Jun 1;73(11):320A-326A. doi: 10.1021/ac012465t. PMID: 11403326.
- Pedersen SK, Harry JL, Sebastian L, Baker J, Traini MD, McCarthy JT, Manoharan A, Wilkins MR, Gooley AA, Righetti PG, Packer NH, Williams KL, Herbert BR. Unseen proteome: mining below the tip of the iceberg to find low abundance and membrane proteins. J Proteome Res. 2003 May-Jun;2(3):303-11. doi: 10.1021/pr025588i. PMID: 12814269.
- Righetti PG, Castagna A, Herbert B, Reymond F, Rossier JS. Prefractionation techniques in proteome analysis. Proteomics. 2003 Aug;3(8):1397-407. doi: 10.1002/pmic.200300472. PMID: 12923764.
- Pedersen SK, Harry JL, Sebastian L, Baker J, Traini MD, McCarthy JT, Manoharan A, Wilkins MR, Gooley AA, Righetti PG, Packer NH, Williams KL, Herbert BR. Unseen proteome: mining below the tip of the iceberg to find low abundance and membrane proteins. J Proteome Res. 2003 May-Jun;2(3):303-11. doi: 10.1021/pr025588i. PMID: 12814269.
- Righetti PG, Castagna A, Herbert B, Candiano G. How to bring the "unseen" proteome to the limelight via electrophoretic pre-fractionation techniques. Biosci Rep. 2005 Feb-Apr;25(1-2):3-17. doi: 10.1007/s10540-005-2844-2. PMID: 16222416.
- Echan LA, Tang HY, Ali-Khan N, Lee K, Speicher DW. Depletion of multiple high-abundance proteins improves protein profiling capacities of human serum and plasma. Proteomics. 2005 Aug;5(13):3292-303. doi: 10.1002/pmic.200401228. PMID: 16052620.
- Huang L, Harvie G, Feitelson JS, Gramatikoff K, Herold DA, Allen DL, Amunngama R, Hagler RA, Pisano MR, Zhang WW, Fang X. Immunoaffinity separation of plasma proteins by IgY microbeads: meeting the needs of proteomic sample preparation and analysis. Proteomics. 2005 Aug;5(13):3314-28. doi: 10.1002/pmic.200401277. PMID: 16041669.
- Zolotarjova, N., Martosella, J., Nicol, G., Bailey, J., Boyes, B.E., Barrett, W.C:, Proteomics 2005, 5, 3304-3313.
- Gianazza E, Giacon P, Astrua-Testori S, Righetti PG. Serum protein analysis on immobilized pH gradients with in situ adsorption of albumin on Dextran Blue. Electrophoresis. 1985; 6: 326-331.
- Zhou M, Lucas DA, Chan KC, Issaq HJ, Petricoin EF 3rd, Liotta LA, Veenstra TD, Conrads TP. An investigation into the human serum "interactome". Electrophoresis. 2004 May;25(9):1289-98. doi: 10.1002/elps.200405866. PMID: 15174051.
- Thulasiraman V, Lin S, Gheorghiu L, Lathrop J, Lomas L, Hammond D, Boschetti E. Reduction of the concentration difference of proteins in biological liquids using a library of combinatorial ligands. Electrophoresis. 2005 Sep;26(18):3561-71. doi: 10.1002/elps.200500147. PMID: 16167368.
- Righetti PG, Castagna A, Antonioli P, Boschetti E. Prefractionation techniques in proteome analysis: the mining tools of the third millennium. Electrophoresis. 2005 Jan;26(2):297-319. doi: 10.1002/elps.200406189. PMID: 15657944.
- Righetti PG, Castagna A, Antonucci F, Piubelli C, Cecconi D, Campostrini N, Rustichelli C, Antonioli P, Zanusso G, Monaco S, Lomas L, Boschetti E. Proteome analysis in the clinical chemistry laboratory: myth or reality? Clin Chim Acta. 2005 Jul 24;357(2):123-39. doi: 10.1016/j.cccn.2005.03.018. PMID: 15970281.
- Merrifield RB. Automated synthesis of peptides. Science. 1965 Oct 8;150(3693):178-85. doi: 10.1126/science.150.3693.178. PMID: 5319951.
- Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. A new type of synthetic peptide library for identifying ligand-binding activity. Nature. 1991 Nov 7;354(6348):82-4. doi: 10.1038/354082a0. Erratum in: Nature 1992 Dec 24-31;360(6406):768. Erratum in: Nature 1992 Jul 30;358(6385):434. PMID: 1944576.
- Furka A, Sebestyén F, Asgedom M, Dibó G. General method for rapid synthesis of multicomponent peptide mixtures. Int J Pept Protein Res. 1991 Jun;37(6):487-93. doi: 10.1111/j.1399-3011.1991.tb00765.x. PMID: 1917305.
- Watts AD, Hunt NH, Hambly BD, Chaudhri G. Separation of tumor necrosis factor alpha isoforms by two-dimensional polyacrylamide gel electrophoresis. Electrophoresis. 1997 Jun;18(7):1086-91. doi: 10.1002/elps.1150180710. PMID: 9237560.
- van Holde KE, Johnson WC, Ho PS. Principles of Physical Biochemistry, Prentice Hall, Upper Saddle River, 1998. 9-11.
- Rabilloud T. Use of thiourea to increase the solubility of membrane proteins in two-dimensional electrophoresis. Electrophoresis. 1998 May;19(5):758-60. doi: 10.1002/elps.1150190526. PMID: 9629911.
- Molloy MP. Two-dimensional electrophoresis of membrane proteins using immobilized pH gradients. Anal Biochem. 2000 Apr 10;280(1):1-10. doi: 10.1006/abio.2000.4514. PMID: 10805514.
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105-32. doi: 10.1016/0022-2836(82)90515-0. PMID: 7108955.
- Hjertén S. Some general aspects of hydrophobic interaction chromatography. J Chromatogr. 1973; 87: 325-331.
- Castagna A, Cecconi D, Sennels L, Rappsilber J, Guerrier L, Fortis F, Boschetti E, Lomas L, Righetti PG. Exploring the hidden human urinary proteome via ligand library beads. J Proteome Res. 2005 Nov-Dec;4(6):1917-30. doi: 10.1021/pr050153r. PMID: 16335936.
- Johnstone A, Thorpe R. Immunochemistry in Practice. Klackwell Sci Publ. Oxford. 1982; 202-232.
- Tanford C. The Hydrophobic Effect: Formation of Micelles and Biological Membranes. John Wiley & Sons, New York, 1973; 100-130.
- Meng R, Gormley M, Bhat VB, Rosenberg A, Quong AA. Low abundance protein enrichment for discovery of candidate plasma protein biomarkers for early detection of breast cancer. J Proteomics. 2011 Dec 21;75(2):366-74. doi: 10.1016/j.jprot.2011.07.030. Epub 2011 Aug 7. PMID: 21851866.
- Fonslow BR, Carvalho PC, Academia K, Freeby S, Xu T, Nakorchevsky A, Paulus A, Yates JR 3rd. Improvements in proteomic metrics of low abundance proteins through proteome equalization using ProteoMiner prior to MudPIT. J Proteome Res. 2011 Aug 5;10(8):3690-700. doi: 10.1021/pr200304u. Epub 2011 Jun 24. PMID: 21702434; PMCID: PMC3161494.
- Peffers MJ, McDermott B, Clegg PD, Riggs CM. Comprehensive protein profiling of synovial fluid in osteoarthritis following protein equalization. Osteoarthritis Cartilage. 2015 Jul;23(7):1204-13. doi: 10.1016/j.joca.2015.03.019. Epub 2015 Mar 26. PMID: 25819577; PMCID: PMC4528073.
- Anderson JR, Phelan MM, Rubio-Martinez LM, Fitzgerald MM, Jones SW, Clegg PD, Peffers MJ. Optimization of Synovial Fluid Collection and Processing for NMR Metabolomics and LC-MS/MS Proteomics. J Proteome Res. 2020 Jul 2;19(7):2585-2597. doi: 10.1021/acs.jproteome.0c00035. Epub 2020 Apr 7. PMID: 32227958; PMCID: PMC7341532.
- Edwards JJ, Tollaksen SL, Anderson NG. Proteins of human urine. III. Identification and two-dimensional electrophoretic map positions of some major urinary proteins. Clin Chem. 1982 Apr;28(4 Pt 2):941-8. PMID: 6804121.
- Pang JX, Ginanni N, Dongre AR, Hefta SA, Opitek GJ. Biomarker discovery in urine by proteomics. J Proteome Res. 2002 Mar-Apr;1(2):161-9. doi: 10.1021/pr015518w. PMID: 12643536.
- Pieper R, Gatlin CL, McGrath AM, Makusky AJ, Mondal M, Seonarain M, Field E, Schatz CR, Estock MA, Ahmed N, Anderson NG, Steiner S. Characterization of the human urinary proteome: a method for high-resolution display of urinary proteins on two-dimensional electrophoresis gels with a yield of nearly 1400 distinct protein spots. Proteomics. 2004 Apr;4(4):1159-74. doi: 10.1002/pmic.200300661. PMID: 15048996.
- Oh J, Pyo JH, Jo EH, Hwang SI, Kang SC, Jung JH, Park EK, Kim SY, Choi JY, Lim J. Establishment of a near-standard two-dimensional human urine proteomic map. Proteomics. 2004 Nov;4(11):3485-97. doi: 10.1002/pmic.200401018. PMID: 15529407.
- Thongboonkerd V, McLeish KR, Arthur JM, Klein JB. Proteomic analysis of normal human urinary proteins isolated by acetone precipitation or ultracentrifugation. Kidney Int. 2002 Oct;62(4):1461-9. doi: 10.1111/j.1523-1755.2002.kid565.x. PMID: 12234320.
- Schaub S, Wilkins J, Weiler T, Sangster K, Rush D, Nickerson P. Urine protein profiling with surface-enhanced laser-desorption/ionization time-of-flight mass spectrometry. Kidney Int. 2004 Jan;65(1):323-32. doi: 10.1111/j.1523-1755.2004.00352.x. PMID: 14675066.
- Tantipaiboonwong P, Sinchaikul S, Sriyam S, Phutrakul S, Chen ST. Different techniques for urinary protein analysis of normal and lung cancer patients. Proteomics. 2005 Mar;5(4):1140-9. doi: 10.1002/pmic.200401143. PMID: 15693063.
- Lafitte D, Dussol B, Andersen S, Vazi A, Dupuy P, Jensen ON, Berland Y, Verdier JM. Optimized preparation of urine samples for two-dimensional electrophoresis and initial application to patient samples. Clin Biochem. 2002 Nov;35(8):581-9. doi: 10.1016/s0009-9120(02)00362-4. PMID: 12498991.
- Smith G, Barratt D, Rowlinson R, Nickson J, Tonge R. Development of a high-throughput method for preparing human urine for two-dimensional electrophoresis. Proteomics. 2005 Jun;5(9):2315-8. doi: 10.1002/pmic.200401267. PMID: 15887186.
- Joo WA, Lee DY, Kim CW. Development of an effective sample preparation method for the proteome analysis of body fluids using 2-D gel electrophoresis. Biosci Biotechnol Biochem. 2003 Jul;67(7):1574-7. doi: 10.1271/bbb.67.1574. PMID: 12913303.
- Spahr CS, Davis MT, McGinley MD, Robinson JH, Bures EJ, Beierle J, Mort J, Courchesne PL, Chen K, Wahl RC, Yu W, Luethy R, Patterson SD. Towards defining the urinary proteome using liquid chromatography-tandem mass spectrometry. I. Profiling an unfractionated tryptic digest. Proteomics. 2001 Jan;1(1):93-107. doi: 10.1002/1615-9861(200101)1:1<93::AID-PROT93>3.0.CO;2-3. PMID: 11680902.
- Davis MT, Spahr CS, McGinley MD, Robinson JH, Bures EJ, Beierle J, Mort J, Yu W, Luethy R, Patterson SD. Towards defining the urinary proteome using liquid chromatography-tandem mass spectrometry. II. Limitations of complex mixture analyses. Proteomics. 2001 Jan;1(1):108-17. doi: 10.1002/1615-9861(200101)1:1<108::AID-PROT108>3.0.CO;2-5. PMID: 11680890.
- Celis JE, Wolf H, Ostergaard M. Bladder squamous cell carcinoma biomarkers derived from proteomics. Electrophoresis. 2000 Jun;21(11):2115-21. doi: 10.1002/1522-2683(20000601)21:11<2115::AID-ELPS2115>3.0.CO;2-K. PMID: 10892722.
- Rasmussen HH, Orntoft TF, Wolf H, Celis JE. Towards a comprehensive database of proteins from the urine of patients with bladder cancer. J Urol. 1996 Jun;155(6):2113-9. PMID: 8618346.
- Williams KM, Williams J, Marshall T. Analysis of Bence Jones proteinuria by high resolution two-dimensional electrophoresis. Electrophoresis. 1998 Jul;19(10):1828-35. doi: 10.1002/elps.1150191047. PMID: 9719566.
- Marshall T, Williams KM. Electrophoretic analysis of Bence Jones proteinuria. Electrophoresis. 1999 Jun;20(7):1307-24. doi: 10.1002/(SICI)1522-2683(19990601)20:7<1307::AID-ELPS1307>3.0.CO;2-P. PMID: 10424453.
- Clark PM, Kricka LJ, Whitehead TP. Pattern of urinary proteins and peptides in patients with rheumatoid arthritis investigated with the Iso-Dalt technique. Clin Chem. 1980 Feb;26(2):201-4. PMID: 7353265.
- Tracy RP, Young DS, Hill HD, Cutsforth GW, Wilson DM. Two-dimensional electrophoresis of urine specimens from patients with renal disease. Appl Theor Electrophor. 1992;3(2):55-65. PMID: 1477113.
- Lapin A, Feigl W. A practicable two-dimensional electrophoresis of urinary proteins as a useful tool in medical diagnosis. Electrophoresis. 1991 Jul-Aug;12(7-8):472-8. doi: 10.1002/elps.1150120704. PMID: 1915240.
- Marshall T, Willams KM, Vesterberg O. Unconcentrated human urinary proteins analysed by high resolution two-dimensional electrophoresis with narrow pH gradients: Preliminary findings after occupational exposure to cadmium. Electrophoresis. 1985; 6: 47-52.
- Gomo ZA, Henderson LO, Myrick JE. High-density lipoprotein apolipoproteins in urine: I. Characterization in normal subjects and in patients with proteinuria. Clin Chem. 1988 Sep;34(9):1775-80. PMID: 3138041.
- Edwards JJ, Anderson NG, Tollaksen SL, von Eschenbach AC, Guevara J Jr. Proteins of human urine. II. Identification by two-dimensional electrophoresis of a new candidate marker for prostatic cancer. Clin Chem. 1982 Jan;28(1):160-3. PMID: 7055903.
- Thongboonkerd V, Malasit P. Renal and urinary proteomics: current applications and challenges. Proteomics. 2005 Mar;5(4):1033-42. doi: 10.1002/pmic.200401012. PMID: 15669002.
- Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002 Nov;1(11):845-67. doi: 10.1074/mcp.r200007-mcp200. Erratum in: Mol Cell Proteomics. 2003 Jan;2(1):50. PMID: 12488461.
- Anderson NL, Polanski M, Pieper R, Gatlin T, Tirumalai RS, Conrads TP, Veenstra TD, Adkins JN, Pounds JG, Fagan R, Lobley A. The human plasma proteome: a nonredundant list developed by combination of four separate sources. Mol Cell Proteomics. 2004 Apr;3(4):311-26. doi: 10.1074/mcp.M300127-MCP200. Epub 2004 Jan 12. PMID: 14718574.
- Omenn GS, States DJ, Adamski M, Blackwell TW, Menon R, Hermjakob H, Apweiler R, Haab BB, Simpson RJ, Eddes JS, Kapp EA, Moritz RL, Chan DW, Rai AJ, Admon A, Aebersold R, Eng J, Hancock WS, Hefta SA, Meyer H, Paik YK, Yoo JS, Ping P, Pounds J, Adkins J, Qian X, Wang R, Wasinger V, Wu CY, Zhao X, Zeng R, Archakov A, Tsugita A, Beer I, Pandey A, Pisano M, Andrews P, Tammen H, Speicher DW, Hanash SM. Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics. 2005 Aug;5(13):3226-45. doi: 10.1002/pmic.200500358. PMID: 16104056.
- Sennels LF, Boschetti E, Lomas L, Righetti PG, Rappsilber J. Mol Cell. Proteomics 2006, submitted.
- Grandi G. Genomics and Proteomics of Vaccines. Wiley Chichester. 2004; 23-44.
- Janson JC. In Janson JC, Rydén L. (Eds.) Protein Purification, Wiley-VCH, New York. 1998; 3-40.
- Simpson RJ. In Simpson, R.J. (Ed.) Purifying Proteins For Proteomics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor. 2004; 17-40.
- Wisniewski R, Boschetti E, Jungbauer A. In Biotechnology and Biopharmaceutical Manufacturing Processing and Preservation. Avis KE, Wu VL. eds; Interpharm Press. 1996; 2: 61-181.
- Ostlund C, Borwell P, Malm B. Process-scale purification from cell culture supernatants: monoclonal antibodies. Dev Biol Stand. 1987;66:367-75. PMID: 3108053.
- Abendroth J, Chatterjee S, Schomburg D. Purification of a D-hydantoinase using a laboratory-scale streamline phenyl column as the initial step. J Chromatogr B Biomed Sci Appl. 2000 Jan 14;737(1-2):187-94. doi: 10.1016/s0378-4347(99)00443-0. PMID: 10681055.
- Gibert S, Bakalara N, Santarelli X. Three-step chromatographic purification procedure for the production of a his-tag recombinant kinesin overexpressed in E. coli. J Chromatogr B Biomed Sci Appl. 2000 Jan 14;737(1-2):143-50. doi: 10.1016/s0378-4347(99)00524-1. PMID: 10681050.
- Brion F, Rogerieux F, Noury P, Migeon B, Flammarion P, Thybaud E, Porcher JM. Two-step purification method of vitellogenin from three teleost fish species: rainbow trout (Oncorhynchus mykiss), gudgeon (Gobio gobio) and chub (Leuciscus cephalus). J Chromatogr B Biomed Sci Appl. 2000 Jan 14;737(1-2):3-12. doi: 10.1016/s0378-4347(99)00406-5. PMID: 10681036.
- Fortis F, Guerrier L, Righetti PG, Antonioli P, Boschetti E. A new approach for the removal of protein impurities from purified biologicals using combinatorial solid-phase ligand libraries. Electrophoresis. 2006 Aug;27(15):3018-27. doi: 10.1002/elps.200500847. PMID: 16807935.
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.




 Save to Mendeley
Save to Mendeley
