Open Journal of Proteomics and Genomics
In silico design of angiotensin-converting enzyme 2 (ACE2) recombinant protein to block the S1 protein pathway of COVID-19 virus
Yaser Ghazi*
Cite this as
Ghazi Y (2020) Mechanistic In silico design of angiotensin-converting enzyme 2 (ACE2) recombinant protein to block the S1 protein pathway of COVID-19 virus. Open J Proteom Genom 5(1): 001-007. DOI: 10.17352/ojpg.000009Coronavirus is a large family of viruses that includes the common cold and the SARS virus. The Chinese corona, or coronavirus, is a new respiratory virus that began in late 2019 and early 2020 in the province of Hubby and Wuhan, China, and became known as COVID-19. The COVID-19 virus genome is a positive single-stranded RNA (ssRNA (+)) and is 29903 nucleotides long, encoding twelve different proteins. One of these proteins is called the S-protein. During the S-protein contamination cycle, it is divided into two subunits, S1 and S2. The subunit S1, which contains the Receptor Binding Protein (RBD), binds directly to the protease domain of the Angiotensin-Converting Enzyme 2 (ACE2) protein and enters the cell through it. In this study first the ACE2 protein sequence extracted from the NCBI site. To convert the protein to an extracellular protein and excrete it out of the cell, the signal peptide sequence was added to the beginning of the recombinant protein and two amino acids, cysteine and asparagine, added to both sides of the signal peptide sequence to create a self-catalyzing process similar to that found in Inteins. The identifiable motif was then incompletely added to both sides of the peptide signal sequence by ACE2 sequences with F-H-L amino acids sequence. Also, amino acids involved in direct interaction between the two subunits of ACE2 protein were inhibited. Dimerization was removed from the amino acid sequence, eventually to improve the lamb The interaction between the two ACE2 proteins designed with the S1 protein virus enhanced the physicochemical properties of the protein designed using the PROTPARAM and GPMAW sites.
Introduction
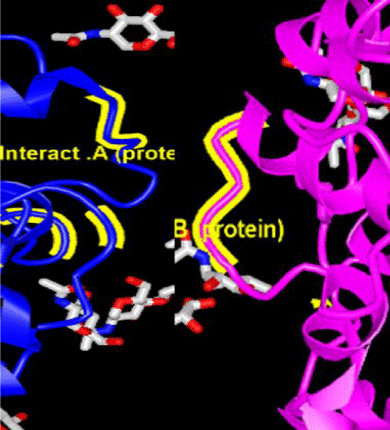
In the vernacular, the coronavirus is called the Wuhan coronavirus. As the number of victims of the Coronavirus virus exceeded 1,000, the World Health Organization (WHO) has chosen the official name of COVID-19 for the disease, which refers to Corona, Virus, Disease and 2019. The COVID-19 virus genome is a positive single-stranded RNA (ssRNA (+)) [1,2], that encodes twelve different proteins. One of these proteins is called S-protein. During the pollution cycle, the S protein is divided into two subunits, S1 and S2. The S1 subunit containing the Receptor Binding Domain (RBD) binds directly to the protease domain of the angiotensin-converting enzyme 2 (ACE2) protein. ACE2 protein acts as a homodimer. The S-protein of the virus also acts as a homotrimer with subunits A, B, and C, and subunit B binds directly to the ACE2 protein.
Initially, such viruses were given the general (and temporary) ncov-2019 name [3-5]. The virus is surrounded by a bubble of lipid molecules that collapse with the presence of soap and water [6-8]. Comparison of the genetic sequence of the virus and other examples of the virus has shown similarities with the SARS and Corona viruses, and therefore may be the primary source of the virus in bats [9,10].
Transmission of the virus from person to person has been confirmed [11]. Coronaviruses are spread primarily through close contact, especially through coughing and sneezing breathing drops of about 2 to 4.5 meters [12,13]. The RNA virus has also been found in fecal samples from infected patients [14]. Studies have shown that the virus can become infected even during the incubation period [15,16]. However, according to the World Health Organization, transmission from asymptomatic individuals is more likely at this time. The main reproductive number (R0) [17] of this virus is estimated to be between 1.4 and 3.9 [18-22]. This means that, if left unchecked, the virus will normally cause 1.4 to 3.9 new infections in each infection. The virus has been shown to be able to transmit at least four people in a chain [23].
The new coronavirus appears to be less dangerous than SARS and is severely present in 15 to 20 percent of cases. Preliminary estimates suggest that the mortality rate for the virus is between 2 and 3 percent [24]. The World Health Organization has published several protocols for testing the disease [25,26]. The standardized method for testing is the polymerase chain reaction of reverse transcription by Real time (rRT-PCR) [27]. This test should be performed using breathing samples prepared from a variety of methods, including a pharyngeal swab or a sputum sample [28]. Results are generally available within a few hours to two days [29,30]. Blood tests can also be used, but this requires two blood samples taken two weeks apart, and the results are not of immediate value [31]. Chinese scientists have been able to isolate a one-way coronavirus and release a nucleic acid sequence so that laboratories around the world can independently develop Polymerase Chain Reaction (PCR) testing laboratories to detect viral infections [32-37].
In this study, an attempt was made to adopt a preventive solution against COVID-19 with the design of a recombinant ACE2 receptor. The ACE2 protein plays a key role in the entry of the virus into the cell. The designed ACE2 protein travels an extracellular pathway with the help of a signal peptide sequence added to the beginning. This causes the COVID-19 virus S1 protein, which interacts directly with the ACE2 protein to enter the cell, to collide with the designed protein before reaching the cell membrane in the extracellular space and block its activity. Amino acids are placed on either side of the signal peptide sequence to act similarly to the exit of intein from the protein sequence, and after the transfer of the designed protein to the extracellular space, the signal peptide sequence leaves the main protein sequence. After the signal peptide sequence is removed from the main sequence, a motif identifiable by ace2 group proteases appears, which is incompletely added to both sides of the signal peptide sequence. Cutting in this motif causes the virus protein to separate from the receptor surface. The incomplete addition of the motif means that no incisions are made before the receptor is transferred out of the membrane.
Materials and methods
First The amino acid sequence of the ACE2 protein was extracted from the NCBI database (http://www.ncbi.nlm.nih.gov). Using bioinformatics software, the protein was analyzed. In order to prevent homodimer formation, part of the protein structure involved in homodimerization was altered. Accordingly, the amino acids involved in the direct interaction between the two subunits were identified and excluded from the initial sequence (Figure 1).
A sequence of proteins involved in a direct connection with the virus’s S-protein was applied without altering the designed protein.
In order to convert membrane protein into a secretory protein using a signal peptide sequence, a number of secretory proteins are predicted by the SignalP site (http://www.cbs.dtu.dk/services/SignalP) and their protected areas by the ClustalO site (http://www.uniprot.org/align/) were identified, a signal peptide sequence was designed for protein secretion outside the cell, and at the beginning of the recombinant protein after the binding motif was added to the S-protein so that the virus could reach the cell membrane and Connect the main protein to the designed protein and block its activity path.
To the two sides of the signal peptide sequence, two amino acids, cysteine and asparagine, were added to perform the same catalytic process as in the proteins, and the signal peptide sequence after transfer. Protein is removed from the cell from the main sequence, which results in a short three-amino acid motif, which is the site for detection and removal by ACE2 proteases and is incompletely added to both sides of the peptide signal sequence by removing the peptide signal sequence appear. By breaking down in this area the virus protein that is connected to the designed protein is released from the protein surface.
Finally, with the help of ProtParam (http://web.expasy.org/protparam) and GPMAW (http://www.alphalyse.com/gpmaw lite.html) sites, the physicochemical properties of the designed protein improved.
Results
The ACE2 protein sequence and the amino acids involved in the interaction between ACE2 and the S1 protein of the COVID-19 virus (red) are shown, also the amino acids involved in the interaction between the two ACE2 subunits for dimerization (yellow), are shown. At the end of the sequence the motif outside the protease is shown.
>pdb|6VW1|A Chain A, Angiotensin-converting enzyme 2
STIEEQAKTFLDKFNHEAEDLFYQSSMNSFSTSAFGPVAFSLGLLLVLPAAFPASWNYNTNITEENVQNMNNAGDKWSAF LKEQSTLAQMYPLQEIQNLTVKLQLQALQQNGSSVLSEDKSKRLNTILNTMSTIYSTGKVCNPDNPQECLLLEPGLNEIMAN SLDYNERLWAWESWRSEVGKQLRPLYEEYVVLKNEMARANHYEDYGDYWRGDYEVNGVDGYDYSRGQLIEDVEHTFEEIK PLYEHLHAYVRAKLMNAYPSYISPIGCLPAHLLGDMWGRFWTNLYSLTVPFGQKPNIDVTDAMVDQAWDAQRIFKEAEKFF VSVGLPNMTQGFWENSMLTDPGNVQKAVCHPTAWDLGKGDFRILMCTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRN GANEGFHEAVGEIMSLSAATPKHLKSIGLLSPDFQEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKDQWMKK WWEMKREIVGVVEPVPHDETYCDPASLFHVSNDYSFIRYYTRTLYQFQFQEALCQAAKHEGPLHKCDISNSTEAGQKL FNMLRLGKSEPWTLALENVVGAKNMNVRPLLNYFEPLFTWLKDQNKNSFVGWSTDWSPYADGSLEVLFQ
2. The addition of two amino acids, cysteine and asparagine, to both sides of the signal peptide sequence to create a self-catalyzing process similar to that found in Inteins.
>pdb|6VW1|A Chain A, Angiotensin-converting enzyme 2
STIEEQAKTFLDKFNHEAEDLFYQSSCMNSFSTSAFGPVAFSLGLLLVLPAAFPNASWNYNTNITEENVQNMNNAGD KWSAFLKEQSTLAQMYPLQEIQNLTVKLQLQALQQNGSSVLSEDKSKRLNTILNTMSTIYSTGKVCNPDNPQECLLL EPGLNEIMANSLDYNERLWAWESWRSEVGKQLRPLYEEYVVLKNEMARANHYEDYGDYWRGDYEVNGVDGYDYSRGQ LIEDVEHTFEEIKPLYEHLHAYVRAKLMNAYPSYISPIGCLPAHLLGDMWGRFWTNLYSLTVPFGQKPNIDVTDAMV DQAWDAQRIFKEAEKFFVSVGLPNMTQGFWENSMLTDPGNVQKAVCHPTAWDLGKGDFRILMCTKVTMDDFLTAHHE MGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKSIGLLSPDFQEDNETEINFLLKQALTIVGTLPF TYMLEKWRWMVFKGEIPKDQWMKKWWEMKREIVGVVEPVPHDETYCDPASLFHVSNDYSFIRYYTRTLYQFQFQEAL CQAAKHEGPLHKCDISNSTEAGQKLFNMLRLGKSEPWTLALENVVGAKNMNVRPLLNYFEPLFTWLKDQNKNSFVGW STDWSPYADGSLEVLFQ
3. Addition of identifiable motif by ACE2 group proteases with incomplete F-H-L amino acids sequence to both sides of signal peptide sequence.
>pdb|6VW1|A Chain A, Angiotensin-converting enzyme 2
STIEEQAKTFLDKFNHEAEDLFYQSSFCMNSFSTSAFGPVAFSLGLLLVLPAAFPNHLASWNYNTNITEENVQNMN NAGDKWSAFLKEQSTLAQMYPLQEIQNLTVKLQLQALQQNGSSVLSEDKSKRLNTILNTMSTIYSTGKVCNPDNPQ ECLLLEPGLNEIMANSLDYNERLWAWESWRSEVGKQLRPLYEEYVVLKNEMARANHYEDYGDYWRGDYEVNGVDGY DYSRGQLIEDVEHTFEEIKPLYEHLHAYVRAKLMNAYPSYISPIGCLPAHLLGDMWGRFWTNLYSLTVPFGQKPNI DVTDAMVDQAWDAQRIFKEAEKFFVSVGLPNMTQGFWENSMLTDPGNVQKAVCHPTAWDLGKGDFRILMCTKVTMD DFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKSIGLLSPDFQEDNETEINFLLKQA LTIVGTLPFTYMLEKWRWMVFKGEIPKDQWMKKWWEMKREIVGVVEPVPHDETYCDPASLFHVSNDYSFIRYYTRT LYQFQFQEALCQAAKHEGPLHKCDISNSTEAGQKLFNMLRLGKSEPWTLALENVVGAKNMNVRPLLNYFEPLFTWL KDQNKNSFVGWSTDWSPYADGSLEVLFQ
F-H-L amino acids form a motif that is detected and cleaved by ACE2 group proteases. Addition of the motif to the protein sequence immediately after the domain binding to the virus causes the incision in this region to separate the domain and associated virus from the surface of the receptor.
4. Adding amino acids to create a disulfide bond between the two sides of the signal peptide sequence.
>pdb|6VW1|A Chain A, Angiotensin-converting enzyme 2
STIEEQAKTFLCKFNHEAEDLFYQCSFCMNSFSTSAFGPVAFSLGLLLVLPAAFPNHLASWNYNTNITCENVQNM CNAGDKWSAFLKEQSTLAQMYPLQEIQNLTVKLQLQALQQNGSSVLSEDKSKRLNTILNTMSTIYSTGKVCNPD NPQECLLLEPGLNEIMANSLDYNERLWAWESWRSEVGKQLRPLYEEYVVLKNEMARANHYEDYGDYWRGDYEVNG VDGYDYSRGQLIEDVEHTFEEIKPLYEHLHAYVRAKLMNAYPSYISPIGCLPAHLLGDMWGRFWTNLYSLTVPFG QKPNIDVTDAMVDQAWDAQRIFKEAEKFFVSVGLPNMTQGFWENSMLTDPGNVQKAVCHPTAWDLGKGDFRILMCT KVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKSIGLLSPDFQEDNETEINF LLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKDQWMKKWWEMKREIVGVVEPVPHDETYCDPASLFHVSNDYSFIR YYTRTLYQFQFQEALCQAAKHEGPLHKCDISNSTEAGQKLFNMLRLGKSEPWTLALENVVGAKNMNVRPLLNYF EPLFTWLKDQNKNSFVGWSTDWSPYADGSLEVLFQ
5. Eliminate amino acids that are involved in direct interaction between the two subunits of ACE2 protein to prevent dimerization.
>pdb|6VW1|A Chain A, Angiotensin-converting enzyme 2
STIEEQAKTFLCKFNHEAEDLFYQCSFCMNSFSTSAFGPVAFSLGLLLVLPAAFPNHLASWNYNTNITCENVQNM CNAGDKWSAFLKEQSTLAQMYPLQEIQNLTVKLQLQALQQNGSSVLSEDKSKRLNTILNTMSTIYSTGKVCNPDN PQECLLLEPGLNEIMANSLDYNERLWAWESWRSEVGKQLRPLYEEYVVLKNEMARANHYEDYGDYWRGDYEVNGV DGYDYSRGQLIEDVEHTFEEIKPLYEHLHAYVRAKLMNAYPSYISPIGCLPAHLLGDMWGRFWTNLYSLTVPFGQ KPNIDVTDAMVDQAWDAQRIFKEAEKFFVSVGLPNMTQGFWENSMLTDPGNVQKAVCHPTAWDLGKGDFRILMC TKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPKHL-SIGLLSPDF---NETEI NFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKDQWMKKWWEMKREIVGVVEPVPHDETYCDPASLFHVSNDYS FIRYYTRTLYQFQFQEALCQAA-GPLHKCDISNSTEAGQKLFNMLRLGKSEPWTLALENVVGAKNMNV-PLLNYF EPLFTWLKDQNKNSFVGWSTDWSPYADGSLEVLFQ
6. According to the ProtParam website, the half-life of ACE2 protein in mammalian bodies is 1.9 hours, which is related to Serine amino acid. Half-life is the time it takes for half of the protein to disappear into the cell after synthesis [38]. According to Table 1, the highest half-life in mammals is that of the amino acid Valine, which is 100 hours. Accordingly, a Valine amino acid was added to the amino acid sequence designed for ACE2 protein.
>pdb|6VW1|A Chain A, Angiotensin-converting enzyme 2
VSTIEEQAKTFLCKFNHEAEDLFYQCSFCMNSFSTSAFGPVAFSLGLLLVLPAAFPNHLASWNYNTNITCENVQNM CNAGDKWSAFLKEQSTLAQMYPLQEIQNLTVKLQLQALQQNGSSVLSEDKSKRLNTILNTMSTIYSTGKVCNPDN PQECLLLEPGLNEIMANSLDYNERLWAWESWRSEVGKQLRPLYEEYVVLKNEMARANHYEDYGDYWRGDYEVNGVD GYDYSRGQLIEDVEHTFEEIKPLYEHLHAYVRAKLMNAYPSYISPIGCLPAHLLGDMWGRFWTNLYSLTVPFGQK PNIDVTDAMVDQAWDAQRIFKEAEKFFVSVGLPNMTQGFWENSMLTDPGNVQKAVCHPTAWDLGKGDFRILMCTK VTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPKHL-SIGLLSPDF---NETEINF LLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKDQWMKKWWEMKREIVGVVEPVPHDETYCDPASLFHVSNDYSFIR YYTRTLYQFQFQEALCQAA-GPLHKCDISNSTEAGQKLFNMLRLGKSEPWTLALENVVGAKNMNV-PLLNYFEPLF TWLKDQNKNSFVGWSTDWSPYADGSLEVLFQ
7. ProtParam then predicted the aliphatic protein index to be 25.25. Aliphatic index is a protein that is defined as the relative volume occupied by aliphatic side chains. Indicates the degree of protein stability from heat-induced denaturation [39], which is calculated according to the following equation:
Aliphatic index = X (Ala) + a * X (Val) + b * (X (Ile) + X (Leu))
In the above relation, x represents the molar percentage of each amino acid, a represents the relative volume of the Valine of the side chain, which is equal to 2.9, and b represents the relative volume of the ratio of Leucine to the Isoleucine of the side chain, which is equal to 3.9. Alanine, Valine, Isoleucine, and Leucine play the most important role in the rate of aliphatic index of a four amino acid protein. Accordingly, to increase the aliphatic index of the recombinant protein, two amino acids, Valine and Isoleucine, were substituted for other amino acids in the extra-protein motif because they had the greatest effect on the above formula.
>pdb|6VW1|A Chain A, Angiotensin-converting enzyme 2
VSTIEEQAKTFLCKFNHEAEDLFYQCSFCMNSFSTSAFGPVAFSLGLLLVLPAAFPNHLASWNYNTNITCENVQN MCNAGDKWSAFLKEQSTLAQMYPLQEIQNLTVKLQLQALQQNGSSVLSEDKSKRLNTILNTMSTIYSTGKVCNPD NPQECLLLEPGLNEIMANSLDYNERLWAWESWRSEVGKQLRPLYEEYVVLKNEMARANHYEDYGDYWRGDYEVNGV DGYDYSRGQLIEDVEHTFEEIKPLYEHLHAYVRAKLMNAYPSYISPIGCLPAHLLGDMWGRFWTNLYSLTVPFGQ KPNIDVTDAMVDQAWDAQRIFKEAEKFFVSVGLPNMTQGFWENSMLTDPGNVQKAVCHPTAWDLGKGDFRILMC TKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPKHL-SIGLLSPDF---NETE INFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKDQWMKKWWEMKREIVGVVEPVPHDETYCDPASLFHVSN DYSFIRYYTRTLYQFQFQEALCQAA-GPLHKCDISNSTEAGQKLFNMLRLGKSEPWTLALENVVGAKNMNV- PLLNYFEPLFTWLKDQNKNSFVGWSTVWSPYADISLEVVFQ
8. Hydrophobic interactions also involve the interpolation of two molecules in which the amino acids Histidine, Phenylalanine, Tryptophan, and Tyrosine are involved [40]. The GPMAW website predicted the ACE2 protein hydrophobicity index to be -0.276. The hydrophobicity index is equal to the amount of energy required to transfer one mole of protein from the hydrophilic medium to the hydrophobic medium. Four amino acids, Histidine, Phenylalanine, Tryptophan, and Tyrosine, replaced other amino acids to increase the hydrophobicity index in a number of situations.
>pdb|6VW1|A Chain A, Angiotensin-converting enzyme 2
VSFIEEQAKTFLCKFYHEAEDLFYQCSFCMNSFSTSAFGPVAFSLGLLLVLPAAFPNHLASWWYNTNITCENV HNMCNAGDKWSAFLKEQSTLAQMYPLQEIQNLTVKLQLQALQQNGSSVLSEDKSKRLNTILNTMSTIYSTGKV CNPDNPQECLLLEPGLNEIMANSLDYNERLWAWESWRSEVGKQLRPLYEEYVVLKNEMARANHYEDYGDYWR GDYEVNGVDGYDYSRGQLIEDVEHTFEEIKPLYEHLHAYVRAKLMNAYPSYISPIGCLPAHLLGDMWGRFWT NLYSLTVPFGQKPNIDVTDAMVDQAWDAQRIFKEAEKFFVSVGLPNMTQGFWENSMLTDPGNVQKAVCHPTA WDLGKGDFRILMCTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPKHL- SIGLLSPDF---NETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKDQWMKKWWEMKREIVGVVEP VPHDETYCDPASLFHVSNDYSFIRYYTRTLYQFQFQEALCQAA-GPLHKCDISNSTEAGQKLFNMLRLGK SEPWTLALENVVGAKNMNV- PLLNYFEPLFTWLKDQNKNSFVGWSTVWSPYADISLEVVFQ
9. ProtParam estimated the protein instability index at 41.73. Instability index is the amount of protein stability inside the test tube [41], which is calculated according to the following equation:
i = L-1
II = (10 / L) * Sum DIWV (x [i] x [i + 1])
i = 1
In the above relation, L represents the length of the protein and the DIWV (x [i] x [i + 1]) expression indicates the value of the instability weight for the diopter located at position i. For stable proteins, the instability index (Ii) is less than 40, and when this index is higher than 40, it means that the protein is unstable. With the changes applied to the protein sequence, the recombinant protein instability index was transferred to less than 39.92.
Discussion
Although many attempts have been made to produce the vaccine, there is still a vaccine or antiviral drug to eradicate it. There is no definitive cure, prevention or response to coronavirus infections in general [42].
Previous operations have been performed on SARS-CAUs because SARS-CAU2 and SARS-CAU both use the ACE2 enzyme to attack human cells [43]. Three vaccination strategies are being considered. In the first solution, researchers hope to make a complete virus vaccine. The use of such a virus, inactive or dead, to achieve a rapid immune response in humans is against a new infection with COVID-19. The second solution is a subcutaneous vaccine to make a vaccine that makes the immune system sensitive to certain subunits of the virus. In the case of SARS-Kav2, such research focuses on spike proteins that help the virus penetrate the ACE2 enzyme. The third solution is nucleic acid vaccines (DNA or RNA vaccines), a new way of vaccinating. Experimental vaccines produced by each of these strategies should be tested for safety and efficacy [44].
In scientific and medical studies, Chinese and Japanese scientists and researchers have found that antiviral drugs such as Lupinavir / Ritonavir have been shown to be useful in treating and preventing the development of coronavirus and even treating the disease. These drugs have been saved, and overall evidence has shown that antiviral drugs have saved the lives of many people with coronavirus [45].
In general, recombinant products, which are associated with genetic manipulation and DNA changes in various organisms, have caused a huge change in the type and variety of pharmaceutical products used, so that today we see the use of high molecular weight pharmaceutical recombinant products instead of small chemical molecules. Protein drugs have a very specific function. Therefore, they will not have an adverse effect on other unrelated biological processes, and in this respect, they have fewer side effects. The rapid growth of biological data has created problems for biologists and biotechnologists to gather, store, and store information in a way that may no longer be possible without the use of new technologies. But in addition to these capabilities, the shadow of ambiguity due to the less predictable effects of this knowledge has led to a challenging future that challenges most social aspects and perhaps most of all genomics.
The function of proteins depends on the spatial structure of the protein, or its third structure, so that many of the defects and dysfunctions of proteins are due to changes in the spatial structure of a protein [46,47]. Extensive data from protein sequencing data have been obtained from modern research methods in molecular biology. The rapid growth of laboratory information on protein sequence and difficult access to protein structure has led to the importance of predicting structure more than ever before [47]. Despite extensive research into the third structure of proteins, the physical basis for the stability of the structure of proteins has not been fully understood [48]. On the other hand, the strong dependence of protein function on its structure has opened new perspectives in the treatment of diseases. Today, the use of information about the third structure of proteins is one of the basic methods in the logical design of drugs [49].
In general, the strategies used to design a vaccine against coronavirus are different from previous strategies used to prevent influenza and colds, and are based more on molecular methods such as DNA, RNA, and recombinant proteins [50]. Among these, the greatest focus is on protein subunits and recombinant proteins [51,52]. The ACE2 protein, as the initiator of the viral infection cycle, can be a good choice for treatment. This protein, which acts as a hemodimer, has been the subject of extensive research because of its high therapeutic potential.
In 2020, Dwight, et al. Tried to block the ACE2 protein with chloroquine and hydroxychloroquine, as well as the antiviral drugs lopinavir and ritonavir, to prevent the virus from attaching to the cell [53]. Mutsuo, et al. Used camovast or naphamostat, commonly used to treat pancreas and intravenous coagulation, to block the pathway of COVID-19 activity into cells [54]. Claudia, et al. Used gene expression profiles including Cancer Genome Atlas, Gene Expression Omnibus and Genotype-Tissue Expression, Gene Ontology and pathway enrichment analysis to evaluate the main functions of ACE2-related genes. Identified 36 potential drugs that could play a key role. Among the interesting drugs for the treatment of COVID-19, Nimesulide, Fluticasone Propionate, Thiabendazole, Photofrin, Didanosine and Flutamide were introduced [55]. Liang-Qin, et al. Using computer screening methods for potential Chinese Herbal Medicine (CHM) for the treatment of COVID-19 by binding the drug molecular device to the hydrolytic enzyme SARS-CoV-2 3CL and converting enzyme Angiotensin II were used as receptors. In this study, they identified six small molecule compounds that have the optimal binding energy to two target proteins. Among the 238 anti-potential COVID-19 plants that have been screened in general, 16 types of CHM contain the most active ingredients, and 5 Candida anti-COVID-19 plants used in high frequency, as well as a pair of medicinal kernels, Forsythiae Fructus-Lonicerae Japonicae Flos, were selected [56]. To block the virus infection cycle, Yuanmei, et al. Designed a lipopeptide fusion inhibitor called IPB02, which showed great potential for inhibiting cell-cell binding [57]. Shuai, et al. Developed a series of EK1-derived lipopeptides previously designed to target the second HR1 on protein S and found that EK1C4 was the most potent fusion inhibitor against protein-mediated protein-mediated SARS-CoV-2 S protein-mediated infection. Viral and viral infection with IC50s is 1.3 and 15.8 nm, about -241 and 149 times stronger than the original EK1 peptide, respectively [58].
However, the use of chemical compounds and base analogues always leave unintended and unforeseen side effects, which are sometimes worse than the original disease for which they were used. However, the use of engineered proteins that have undergone engineering manipulations, in addition to not having such side effects, do not stimulate the immune system against the designed protein and have a higher sensitivity and specificity.
- Gobalenya AE, Baker SC, Baric RS, Groot JR, Drosten C, et al. (2020) The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology 5: 536-544. Link: https://go.nature.com/2WNCCKs
- Coronavirus disease named Covid-19. BBC News Online. Link: https://bit.ly/2ZQyong
- Surveillance case definitions for human infection with novel coronavirus (nCoV): interim guidance v1, January 2020 (Report). World Health Organization. 2020. Link: https://bit.ly/2Bs1u2U
- Healthcare Professionals: Frequently Asked Questions and Answers. U.S. Centers for Disease Control and Prevention (CDC). Link: https://bit.ly/32JblfX
- About Novel Coronavirus (2019-nCoV) . U.S. Centers for Disease Control and Prevention (CDC). Link: https://bit.ly/32L64V0
- How Coronavirus Hijacks Your Cells. Link: https://nyti.ms/3hxr3iv
- Zhou P, Yang XL, Shi ZL (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579: 270-273. Link: https://go.nature.com/2CFn9oR
- Zhou P, Yang XL, Shi ZL (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579: 270-273. Link: https://go.nature.com/2CFn9oR
- The 2019-new coronavirus epidemic: evidence for virus evolution. Link: https://bit.ly/2WQkcbR
- Why snakes probably aren’t spreading the new China virus. Nature. Link: https://go.nature.com/2ZT4pLx
- China confirms human-to-human transmission of new coronavirus. Canadian Broadcasting Corporation. Link: https://bit.ly/2OOQdwD
- How does coronavirus spread?. NBC News. Link: https://bit.ly/2OSHadS
- Transmission of Novel Coronavirus (2019-nCoV) | CDC. www.cdc.gov. Link: https://bit.ly/2ZU8K0E
- Holshue Michelle L, DeBolt C, Lindquist S, Lofy Kathy H, Wiesman J, et al. (2020) First Case of 2019 Novel Coronavirus in the United States. New England Journal of Medicine 382: 929-936. Link: https://bit.ly/2X57iHl
- Rothe C, Schunk M, Sothmann P, Bretzel G, Froeschl G, Wallrauch C, et al. (2020) Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany. The New England Journal of Medicine 382: 970-971. Link: https://bit.ly/3jzlwd2
- There's no doubt. Top US infectious disease doctor says Wuhan coronavirus can spread even when people have no symptoms". CNN. Link: https://cnn.it/32MXw07
- Milligan Gregg N, Barrett ADT (2015) Vaccinology: an essential guide. Chichester, West Sussex: Wiley Blackwell. 310. Link: https://bit.ly/2OQ211z
- The Deceptively Simple Number Sparking Coronavirus Fears. Link: https://bit.ly/2BngKhm
- Qun L, Xuhua G, Peng W, Xiaoye W (2020) Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus–Infected Pneumonia. New England Journal of Medicine 382: 1199-1207. Link: https://bit.ly/2ZXtJjz
- Julien R, Christian LA (2020) Pattern of early human-to-human transmission of Wuhan 2019 novel coronavirus (2019-nCoV), December 2019 to January 2020. Euro Surveill 25: 2000058. Link: https://bit.ly/2EfOdM1
- Tao L, Jianxiong H, Min K, Lifeng L (2020) Transmission dynamics of 2019 novel coronavirus (2019-nCoV). bioRxiv. Link: https://bit.ly/2BpgBtP
- Read Jonathan M, Bridgen Jessica RE, Cummings Derek AT, Ho Antonia, Jewell Chris P (2020) Novel coronavirus 2019-nCoV: early estimation of epidemiological parameters and epidemic predictions. MedRxiv. Link: https://bit.ly/2CIQ4Zd
- Saey TH (2020) How the new coronavirus stacks up against SARS and MERS. Science News. Link: https://bit.ly/3jy3Nmk
- Wuhan Coronavirus Death Rate - Worldometer. www.worldometers.info. Link: https://bit.ly/3fUPq9o
- Schirring L (2020) Japan has 1st novel coronavirus case; China reports another death. CIDRAP. Archived from the original on 20 January 2020. Retrieved 16 January 2020.
- Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases: Interim guidance. Link: https://bit.ly/2ZTzkXP
- 2019 Novel Coronavirus (2019-nCoV) Situation Summary. 30 January 2020. Link: https://bit.ly/2CFA6yX
- Real-Time RT-PCR Panel for Detection 2019-nCoV. Link: https://bit.ly/2E3mvSj
- Curetis Group Company Ares Genetics and BGI Group Collaborate to Offer Next-Generation Sequencing and PCR-based Coronavirus (2019-nCoV) Testing in Europe. GlobeNewswire News Room (2020). Link: https://bit.ly/32Oc1R1
- Brueck H (2020) There's only one way to know if you have the coronavirus, and it involves machines full of spit and mucus. Business Insider. Link: https://bit.ly/2WLZb2c
- Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases. Link: https://bit.ly/39ow1Li
- "Undiagnosed pneumonia – China (HU) (01): wildlife sales, market closed, RFI Archive Number: 20200102.6866757". Pro-MED-mail. International Society for Infectious Diseases. Link: https://bit.ly/2D1oAxw
- Cohen J, Normile D (2020) New SARS-like virus in China triggers alarm (PDF). Science 367: 234-235. Link: https://bit.ly/3fYOy3u
- Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1, complete genome. NCBI. Link: https://bit.ly/39kHT17
- Severe acute respiratory syndrome coronavirus 2 data hub. NCBI. Link: https://bit.ly/3jwjtq0
- SARS-CoV-2 (Severe acute respiratory syndrome coronavirus 2) Sequences. NCBI. Link: https://bit.ly/30y2g6M
- Genomic epidemiology of SARS-CoV2. GISAID. Link: https://bit.ly/30IOM8F
- Ciechanover A, Schwartz AL (1989) How are substrates recognized by the ubiquitin-mediated proteolytic system? Trends Biochem Sci 14: 483-488. Link: https://bit.ly/3huV1ni
- Ikai AJ (1980) Thermostability and aliphatic index of globular proteins. J Biochem 88: 1895-1898. Link: https://bit.ly/2CWtUCC
- Kamram HM (2010) Computational Analysis of Protein Ligand Interaction, university of York.
- Guruprasad K, Reddy BVB, Pandit MW (1990) Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng 4: 155-161. Link: https://bit.ly/39ofRBD
- Hui DS, I Azhar E, Madani TA, Ntoumi F, Kock R, et al. (2020) The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health – The latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis 91: 264-266. Link: https://bit.ly/3fTQWbN
- Cascella M, Rajnik M, Cuomo A, Dulebohn SC, Di Napoli R (2020) Features, Evaluation and Treatment Coronavirus (COVID-19) . StatPearls. Link: https://bit.ly/2WYXe1t
- Chen W, Strych U, Hotez PJ, Bottazzi ME (2020) The SARS-CoV-2 Vaccine Pipeline: an Overview. Current Tropical Medicine Reports 7: 61-64. Link: https://bit.ly/2ZTAAdv
- Reuters: us china health abbvie hiv. Link: https://reut.rs/2CXEB7N
- Lehrman SR (1990) Protein structure. Bioprocess Technol 7: 9-38. Link: https://bit.ly/39l78jO
- Alberts B (2002) Molecular Biology of the Cell, Garland Science. Link: https://bit.ly/3eVinAL
- Blundell TL, Sibanda BL, Montalvao RW, Brewerton S, Chelliah V, et al. (2006) Structural biology and bioinformatics in drug design: opportunities and challenges for target identification and lead discovery. Philos Trans R Soc Lond B Biol Sci 1467: 413-423. Link: https://bit.ly/3eReZ9N
- Edwards YJ, Cottage A (2001) Prediction of protein structure and function by using bioinformatics. Methods Mol Biol 175: 341-375. Link: https://bit.ly/3fPd4UC
- Thanh Le T, Andreadakis Z, Kumar A, Gómez Román R, Tollefsen S, et al. (2020) The COVID-19 vaccine development landscape. Nature Reviews Drug Discovery 19: 305-306. Link: https://go.nature.com/3hvlnpa
- Draft landscape of COVID 19 candidate vaccines". World Health Organization. Link: https://bit.ly/2P2ennD
- "COVID-19 vaccine development pipeline (Refresh URL to update)". Vaccine Centre, London School of Hygiene and Tropical Medicine. Link: https://bit.ly/2Bo1Jfg
- Dwight LM, Ariane S, Ulrike S, Stefan L, Cord N (2020) Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacol Res 157: 104859. Link: https://bit.ly/32OENRU
- Mutsuo Y , Hidekazu N , Xue D, Akiko K , Ryoichi N (2020) Protease Inhibitors: Candidate Drugs to Inhibit Severe Acute Respiratory Syndrome Coronavirus 2 Replication. Tohoku J Exp Med 251: 27-30. Link: https://bit.ly/32MF3kb
- Claudia C , Gloria B , Isabella C (2020) In Silico Discovery of Candidate Drugs against Covid-19. Viruses 12: 404. Link: https://bit.ly/3jxerts
- Liang-Qin G, Jing X, Shao-Dong C (2020) In Silico Screening of Potential Chinese Herbal Medicine Against COVID-19 by Targeting SARS-CoV-2 3CLpro and Angiotensin Converting Enzyme II Using Molecular Docking. Chin J Integr Med 26: 527-532. Link: https://bit.ly/2ZQWmyw
- Yuanmei Z, Danwei Y , Hongxia Y , Huihui C , Yuxian H (2020) Design of Potent Membrane Fusion Inhibitors against SARS-CoV-2, an Emerging Coronavirus with High Fusogenic Activity. J Virol 94: e00635-20. Link: https://bit.ly/2CJnOFE
- Shuai X , Meiqin L , Chao W, Wei X , Qiaoshuai L, et al. (2020) Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res 30: 343-355. Link: https://go.nature.com/3jwW33H
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
