Open Journal of Proteomics and Genomics
Recent advances in understanding cross-talk between Bile Acids and Gut Microbiota
Cheng-cheng Feng1,2, Ai-hua Zhang2, Jian-hua Miao1, Hui Sun2, Ying Han2, Guang-li Yan2, Fang-fang Wu1 and Xi-jun Wang1,2*
Cite this as
Feng CC, Zhang AH, Miao JH, Sun H, Wang XJ, et al. (2018) Recent advances in understanding cross-talk between Bile Acids and Gut Microbiota. Open J Proteom Genom 3(1): 024-034. DOI: 10.17352/ojpg.000007Bile acid (BA) plays an important role in the absorption and translocation of fat and fat-soluble vitamins. In addition, it can also act as a signaling molecule to influence the energy metabolism of organisms, glucose metabolism, and the development of liver and intestinal diseases by activating receptor. Gut microbiota participates in the metabolism and transport of BA, which changes the BA associated with the occurrence and development of a variety of diseases. This is achieved through a variety of regulatory processes and is intrinsically linked to host physiology. In recent years, many scholars have used 16S rRNA gene sequencing in conjunction with serum, urine, and fecal metabolomics methods to study the mechanisms underlying the occurrence and development of disease associated with BA and gut microbiota, or to evaluate the protective action of drugs on the metabolic phenotype in rats with gut microbiota disorder. On the one hand, the gut microbiota regulates BA by activating receptors such as FXR, TGR5, and FGF15, and can regulate BA synthesis through enzyme reaction. In addition, gut microbiota can effectively hydrolyze bound parasites or heterogenous organisms that have been cleared by BA. On the other hand, BA can alter the composition of the gut microbiota by inhibiting the growth of bacteria in the intestine. These studies provide new ideas for further elucidating the relationship between gut microbiota and BA and treatment for related disease.
Introduction
In modern scientific research, we have learned that there are hundreds of billions of microorganisms in living organisms. However, how these microorganisms coexist with the host is now a hot research topic in the field of science. The gut microbiota can produce a variety of secondary metabolites that accumulate in the blood and have a systemic effect on the host [1]. Instability or dysbiosis of the gut microbiota and its metabolites may be related to the causes of immune, metabolic and neurological diseases [2-4]. Therefore, research on gut microbiota has become a new direction for a series of diseases prevention and control.
The host genome and the intestinal microflora as important energy substrates and signal factors collectively produce a good deal of metabolites including bile acid (BA), choline, neurotransmitters, short chain fatty acid (SCFAs). The crosstalk between gut microbiota-small molecule metabolite-host regulates metabolic phenotypes and the immune system, and affects risk targets for disease and the response to treatment [5,6]. BA is currently receiving increasing attention in a variety of endogenous metabolites produced by co-metabolism of the host-gut microbiota. BA is an important substance in bile. It regulates the digestion and absorption of lipids, prevents the formation of gall-stone, and regulates the intestinal endometrial homeostasis through the enterohepatic circulation. Moreover, BA plays the role of nutrient signaling hormones by combining with receptors that regulate the expression of genes involved in BA synthesis and transport by encoding enzymes and proteins [7], for instance, farnesoid X receptor (FXR), Takeda-G-protein-receptor-5(TGR5), pregnane X receptor (PXR), vitamin D receptor (VDR), and sphingosine-1-phosphate receptor 2 (S1P2) [6,8-10]. As an important signaling molecule, BA is involved in regulating the physiological activities of the host, which is also closely related to many metabolic and immune diseases in the body, such as obesity, type 2 diabetes, inflammation, liver and intestinal disease [5,11].
Even if the concentration of these small molecules produced in the host blood can reach the same level as the drug, which microorganisms are clearly controlling this metabolic process to regulate the production of these secondary metabolites, and thus affecting the host still needs further research [1]. At present, studies have confirmed that the metabolism of BA is affected by the metabolism of gut microbiota that is an important factor affecting BA absorption and signaling pathways, thus acting on host metabolism, such as obesity, lipid metabolism, diabetes, liver disease, intestinal mucosal function, colon cancer and cardiovascular are related to BA metabolism and BA signal regulation [711,12-18]. Therefore, this paper reviews the understanding cross-talk between bile acids and gut microbiota.
Enterohepatic circulation of bile acid
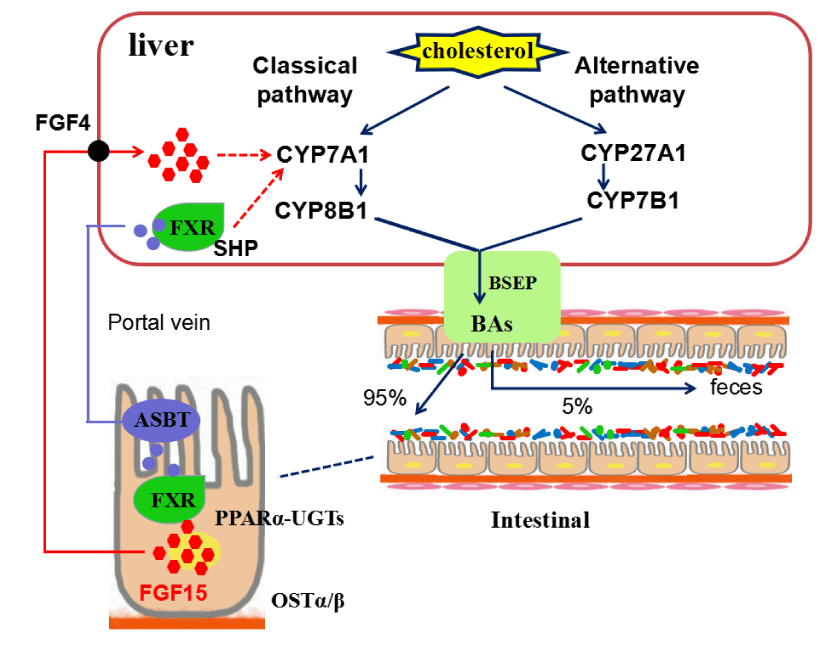
BA is an amphiphilic steroid molecule synthesized from cholesterol [19]. There are two ways to synthesize bile acid. The main pathway for the bile acid synthesis is mediated by Cholesterol 7 alpha-hydroxylase (Cyp7a1) that is a rate-limiting enzyme, and a small part (3%~18%) of the BA pool is synthesized through alternative pathway catalyzed by sterol 27-hydroxylase (Cyp27a1) and oxysterol and steroid 7-alpha-hydroxylase (Cyp7b1) [20], which is shown in figure 1. What significant to realize is that bile acid metabolites between rat/mice and humans differ due to rodent specificity; the conversion of hydrophilic muricholic acid (antagonists of FXR) to hydrophobic chenodeoxycholic acid (agonists of FXR) [21].
In the liver, primary BA is conjugated to glycine (taurine in rat/mice) at the C24 position by BA-CoA amino acid N-acetyltransferase (BAAT) and BA-CoA synthase (BACS). Furthermore, bile acid can also be glucuronidated by UDP -glucuronosyltransferases, or sulfotransferase by SULT2A1-mainly in human-or SULT2A9-mainly in rat/mice. And then, bile acid is secreted into the bile canaliculi with the help of the BA transporters MDR1A and MRP2 and the bile salt export pump (BSEP). Bile acid flows through the biliary tree along with the bile into the gallbladder. enteroendocrine I cells secrete cholecystokinin (CCK) to induce gallbladder contraction after eating, which leads to bile enters the duodenum [22].
In the intestine, BA activates pancreatic lipase and forms micelles containing dietary fat and lipophilic vitamins (A, d, E and K). The gut microbiota converts primary BA to secondary BA. The vast majority of bile acid leaves the liver in the conjugated form, and they are uncoupled and further metabolized by gut microbiota in the small intestine [23-25]. In the intestinal cells at the end of the ileum, 95% of intestinal BA are actively reabsorbed through the apical-sodium-dependent BA transporter (ASBT/SLC10A2) and secreted by the heterodimeric organic solute transporters α and β (OST α/β) at the basolateral membrane. The absorbed BA return to the liver through portal veins and the superior mesenteric, in which active transporters in the sinusoidal membrane of hepatocytes(OATP, OAT,mEH, NTCP) clear them [26]. Unabsorbed 5% intestinal BA can be unbound by the intestinal microbiota and lost into feces or passively reabsorbed in the colon. The enterohepatic circulatory system is a highly effective physiological system for the human which can not only maintain the energy balance of the organism, but also facilitate the absorption and digestion of nutrients [27].
Signal regulation of bile acid synthesis, transport and metabolism
BA not only improves the absorption of fat-soluble nutrients, but also regulates many metabolic processes, including lipids, glucose, and energy homeostasis, by means of specific diacid-activated nuclear and membrane-bound receptors. These receptors regulate the expression of genes encoding enzymes and proteins to control the synthesis and transport of bile acids [28]. FXR is a important regulator of bile acid enterohepatic circulation and homeostasis, and it has high expression levels both in the intestine liver [29]. FXR modulates the expression of genes taked part in the transport and synthesis of bile acid and can be combined with a variety of highly effective endogenous BA and their conjugates. Accumulation of a mass of of toxic bile acids may lead to liver damage and inflammation and cholestatic liver disease. Therefore, BA synthesis is strictly regulated by BA that return to the liver cells from the ileum to restrain the transcription of the CYP8B1 gene and the Cyp7a1 gene through two mechanisms. In the liver BA binds to FXR, which in turn activates Shp to then inhibit their own synthesis in the liver and uptake in the ileum. In the intestine, BA activates FXR to induce fibroblast growth factor 15 (FGF15) which circulates to the liver to inhibit Cyp7a1 gene and CYP8B1 gene transcription by activating membrane FGF receptor 4 signaling [30-34]. Recent studies have shown that the MAF bZIP transcription factor G (MAFG)was the target of FXR. It provides negative feedback regulation of enterohepatic by FXR for bile acid synthesis and is a key transcriptional inhibitor of bile acid synthesis and metabolism [35]. FXR in the ileum controls the BA absorption from the small intestine to the portal vein by inducing ASBT (SLC10A2) that transports BA into epithelial cells, bile acid binding protein (I-BABP) that transports in the BA across the cell, and OSTα/β (SLC51A/B) that transports BA into the portal blood [37]. But when the ileal FXR signal is selectively activated, the mRNA levels of these genes increase [37]. There was a study that had established a method combining liquid chromatography mass spectrometry (LC-MS) with ileal and hepatic RNA-Seq analysis and complementary histopathological assessment, which demonstrated that TCDD increases levels of secondary bile acids in serum and disrupts feedback inhibition induced by bile acids, and conjugated secondary bile acid levels in the liver including GDCA and TLCA by inhibiting the FXR signaling pathway [38]. Increased levels of liver TLCA and other hydrophobic bile acid may cause hepatotoxicity by inducing bile duct proliferation and impairing bile flow that altered primary and secondary bile acid profiles. Bile acids and FXR can also affect sepsis by controlling NLRP3 inflammasome. Bile acids, the danger-associated molecular patterns (DAMPs), can activate both signal 1 and 2 of the NLRP3 inflammasome in inflammatory macrophages. The NLRP3 inflammasome is negatively regulated by FXR through physical interaction between caspase 1 and NLRP3 [39].
Fibroblast growth factor 15 (FGF15), an enterokintic, can participate in many metabolic processes in the organism. Studies had shown that ileal FXR activation promoted FGF15 (human FGF15 orthologs) derived from the intestine and can act on the liver gene transcription in mice [40]. What had been found was that secondary bile acid (TDCA) played the very important role in increasing ileal Fgf15 gene mRNA levels in mouse experiments [41].
Takeda G-protein coupled receptor-5 (TGR5), a G protein-coupled receptor, is an important factor in maintaining glucose homeostasis [42]. In contrast to FXR, TRG5 is mainly activated by some secondary bile acid. BA activates PKA kinase through the TGR5 receptor and directly phosphorylates the serine at position 291 of NLRP3, leading to ubiquitination of NLRP3, which plays an important role in inhibiting the activation of NLRP3 inflammatory bodies. At the same time, this molecular mechanism may also be involved in the pathogenesis of inflammatory diseases caused by the excessive activation of certain NLRP3 inflammatory bodies [43]. TGR5 signaling induces GLP-1 secretion from intestinal L cells to enhance pancreatic and liver function, and increase glucose tolerance in obese mice [44,45]. TGR5 also responds to ligands for bile acids in the liver [46]. When TGR5 is activated, circulating AMP and associated downstream effects increase [47]. Feeding Colesevelam changes the effect of fecal bile acid on TGR5 signaling, which is beneficial to increase the expression of GLP-1 in the colon and increase its level in the portal vein blood, and may help to improve cholestatic liver injury in Mdr2-/- mice [48]. Recently, TGR5 was identified as associated with rapid induction of hepatic steatosis and bile acid synthesis in mice [49]. Studies havd also shown that TGR5 promoted browning of adipose tissue [50]. In addition, the latest reports indicated that bile acid mediated the expression of TGR5 and FXR receptor signals, which reduced the incidence of cardiovascular disease [51].
Regulation of gut microbiota on bile acid
BA is a digestive juice secreted by the liver to help break down fat. The gut microbiota can mediate the conversion of primary BA to secondary. Blood from the gut passes through the liver and brings in metabolites from gut bacteria, which account for 70% of the liver’s blood supply. The liver bile acid balance is affected by intestinal bacterial metabolism [52].
Liver enzymes and bacterial enzymes are involved in a wide variety of bile acid synthesis. Microorganisms involved in bile acid synthesis increase the diversity of bile acid by dehydroxylation of conjugate dehydrogenation and acidification reaction. Bacteroides and Lactobacillus increase the expression of bile salt hydrolase (BSH) by catalyzing the separation of early bile salts to release primary bile acid [53]. In addition, some bacteria containing hydroxysteroid dehydrogenase can mediate bile salt oxidation and isomerization [54].
Gut microbiota regulates bile acid synthesis through enzyme reactions. Antibiotic treatment inhibits the expression of FGF15 and upregulation of CYP7A1. What has also been found was that the expression levels of some enzymes, including CYP7A1 and CYP27A 1, were reduced in sterilized mice compared to normal mice [55]. Antibiotic treatment may change the structure of the intestinal microflora, thereby altering the bile acid composition in the intestine to inhibit FXR signaling in the ileum. Elimination of commensal flora with non-absorbable antibiotics decreased the expression of Cyp7a1 in the liver of mice and inhibited alcoholic liver disease, suggesting that intestinal bacteria modulate bile acid synthesis, which is consistent with the above findings [56]. The BSH released by the gut bacteria acts on the amide bond of the bile salt and reacts to produce glycine or taurine as well as free bile acid; the gut bacteria can further catalyze the dehydroxylation of free cholic acid. It has been reported that BSH is commonly found in Bacteroides, Clostridium, Bifidobacterium, Lactobacillus and Enterococcus [57]. However, different strains have different BSH activities. For example, L. helveticus and L. fermentum (yeast) can only dissociate taurine bile salts and cannot dissociate glycine bile salts. BSH in microorganisms may have the ability to recognize steroids and amino acids. In addition, it has been reported that the addition of BSH-active bacterias can effectively reduce serum and cholesterol levels in mice and dogs [58]. HFD-induced mice treated with antibiotics or tempol modified their microbiota resulting in a reduction in poor metabolic phenotypes, which was due to decreased activity of BSH and decreased levels of Lactobacillus. A decrease in BSH leads to an increase in taurine-beta-murichic acid (T-beta-MCA) levels that is a potent FXR antagonist and a substrate for BSH [59]. A study further showed that antibiotics interfere with intestinal microflora, which can increase the content of FXR receptor antagonist (T-β-MCA) and thus inhibit the FXR receptor [60].
BA plays a key role in the pathological development of inflammatory bowel disease (IBD). The signal of intestinal peroxisome proliferator-activated receptor alpha PPARα-UDP-glucuronosyltransferase (PPARα-UGTs) is an important determinant of bile acid homeostasis. Colitis induced by dextran sulfate sodium (DSS) caused accumulation of BA in inflamed colon tissue by activating PPAR-UGTs pathway. UGT accelerated the elimination of metabolism of BA, thereby reducing their intracellular levels in the small intestine. Decreased intracellular BA inhibited FXR-FGF15 signaling, leading to upregulation of liver CYP71, there by promoting new bile synthesis. The researchers demonstrated that knockout of PPAR and treatment with recombinant FGF19 (rodent FGF15) significantly ameliorated colitis induced by DSS [61].
Intestinal dysbacteriosis can alter bile acid in the intestine and can also be associated with many diseases [62]. At present, the criticality of gut microbiota in host homeostasis and in the pathogenesis of intestinal barrier dysfunction and metabolic diseases such as cancer, type 2 diabetes (T2D), nonalcoholic fatty liver and even mental disorders is recognized [63-65]. There is growing evidence that intestinal flora can exacerbate the development of NAFLD in various animal and human [66,67].
Host-microbe metabolic interactions have a significant effect on heterologous metabolism [68]. Clayton et al. had demonstrated that metabolic interactions in the intestinal microflora led to alteration of major heterogeneous organisms metabolize cytochrome enzymes and cholic acid metabolites [69]. In another study, the gut microbiota was identified to promote the positive effects of cold on obesity through mechanisms involved in producing conjugated bile acid and AMPK activation [70]. Cold exposure can also result in changes in the bile acid composition, including an increase in secondary BA, and the transfer of gut microbiota reduces diet-induced obesity in sterile mice [71]. A study had shown that morphine-induced intestinal metabonomics changes gradually with different changes in individual metabolites. Differential changes in intestinal metabonomics may reflect changes in the gut microbiota, which was conducive to the host’s response after morphine treatment. The cross-correlation between the gut microbiome and the metabolome suggests an association between bacterial communities and functional metabolites [72].
Recently, researchers have discovered that bile acids that can be altered by bacteria that live in the large intestine can inhibit the growth of C. difficile, a harmful bacteria that can cause pain or even death in the body. The use of special antibiotics can cause high-risk bacterial infections in the body. The investigators observed differences in intestinal contents between mice before and after treatment with a variety of different antibiotics, identified 26 differential primary bile acids and secondary bile acids, and determined their concentrations. The researchers found that in a healthy intestinal environment, normal intestinal bacteria produce secondary bile acids to block the growth of C. difficile; and when these bacteria do not exist after antibiotic therapy, these C. difficile will grow quickly 55 . The study of how the spores of C. difficile interact with the microbial community provides a new idea for the mechanism of gut microbiota regulation of BA. With further research, new therapies for Clostridium difficile can be developed and proposed reasonable advice on the use of antibiotics.
In recent years, beige fat has attracted more and more attention in the field of obesity research. The liver and gut microbiota promote the conversion of white fat to beige fat by secreting bile acids. In the blood, BA accumulation and its interaction with the TGR5 receptor can alter the metabolic function of white fat cells. Not only is the color of the cells affected, but BA also increased the number of mitochondria in the new fat cells, indicating that these newly transformed beige fat cells have higher levels of energy metabolism, and BA analogs also cause lipolysis. Letting fat cells use fatty acids as their primary fuel source is the first step in fat degradation in the body. “The role of bile acids is not only to promote digestion, but when the concentration of bile acids in the blood reaches a certain level, fat burning and calorie production in humans/animals will increase, thereby better maintaining body temperature and promoting metabolism. With the direct link between bile acids and fat loss, people have a new intervention strategy for obesity treatment [73,74].
Studies by a research group have shown that metformin increases the level of GUDCA by inhibiting the BSH activity produced by Bacteroides fragilis. Further mechanistic studies have revealed that GUDCA is an endogenous antagonist of human farnesoid X receptor FXR and has potential therapeutic effects on type 2 diabetes. Metformin exerts a hypoglycemic effect through the intestinal tract Bacteroides-Bile acid GUDCA-intestinal FXR metabolic axis. This study analyzed the remodeling effect of metformin on intestinal flora and its metabolites; explored the target of colonic flora in the host and its function; and suggested that bile acid GUDCA and intestinal FXR can be used as a treatment for obesity. A new target for related metabolic diseases [75].
Increased amounts of certain intestinal metabolites such as DCA, TCA, TLCA and TCNCA promote the development of liver tumors [76]. High fat diet (HFD) feeding leads to changes in the gut microbiota of mice. The microbiome promotes cancer by emitting certain signals that alter the microenvironment, and liver stellate cells are important sensors for these signals. The researchers also identified a specific intestinal bacterial metabolite: carcinogen deoxycholic acid (DCA) is a trigger for SASP. Inhibition of DCA production can inhibit liver cancer in obese mice, and treatment with antibiotics in mice with a high-fat diet containing DCA can lead to a surge in liver cancer. It also promotes the formation of colon and esophageal cancer, especially in the case of obesity [77,78]. An increase in the amount of bile acids, such as DCA, TCA, TLCA and TCDCA, promotes the development of liver tumors [79,80].
In the gastrointestinal tract, destruction of the enterohepatic circulation of hepatic bile acids is a cause of inflammation in the liver. Intestinal symbiotic bacterial metabolites play an important role in regulating gastrointestinal barrier function through pregnane X receptor (PXR)-dependent TLR4 signaling [81]. BA promotes the expression of pro-inflammatory cytokines such as IL1β and TNF, thereby inhibiting the transcription of FXR target genes [82]. The down-regulation results in the transfer of bile acids from the intestinal cells back to the portal vein via the IBABP and OSTαOMSβ complex. Therefore, bile acids are accumulated in the intestinal mucosa [83]. Patients with liver diseases such as fibrosis, HCC and cirrhosis frequently manifest intestinal disorders characterized by BSH rich bacteria, pro inflammatory, a major elevation of aerobic enterobacteria that Can promote the production of secondary bile acids, including the genera Enterococcus, Enterobacter and Clostridium [84]. In contrast, In contrast, patients with intestinal diseases typically exhibit intestinal disorders characterized by decreased microbial diversity and reduced bacterial abundance at the phylum Firmicutes. For example, Faecalibacterium prausnitzii causes a decrease in secondary bile acid levels and an increase in conjugated bile acids in the intestine [85-87]. These microbial populations induce bile acid profile changes, which may further result in decreased FXR activity, increased bile acid content in hepatocytes and intestinal mucosa, and decreased expression of liver BSEP, OSTα OSTβ complexes in the liver and intestinal [83,88]. Moreover, the use of probiotics to alter gut microbiota characteristics increases excretion of bile acids in feces and promotes the synthesis of bile acids in the liver of mice, the mechanism of which involved reducing the reabsorption of bile acids from the ileum and inhibiting the FXR-FGF15 axis of the intestine-liver, suggesting that dysplastic probiotics can reverse abnormal bile acid metabolism [89].
Gut microbiota participates in the regulation of nutrient hormone signaling processes regulated by bile acids [90]. Microbiota can regulate BA by activating FXR [91]. As previously reported, reduction of FXR signaling may be due to inhibition of Abcb11. Abcb11 has been identified as a target gene for FXR, which delivers newly synthesized primary bile acids from liver to the bile canaliculi [92]. This is detrimental to bile collection in the gallbladder, which leads to accumulation of bile in the liver cells and eventually spillovers to sinusoidal blood. Acarbose consumes Bacteroides in the gut microbiota and increases the relative abundance of bifidobacteria and lactobacilli, which alters the relative abundance of microbial genes involved in bile acid metabolism, and thereby changes the bile acid profile in plasma and feces. The level of unconjugated BA in the plasma of patients with type 2 diabetes (T2D) who have not receive acarbose treatment, the ratio between primary BA and secondary BA increases after treatment, which enhances FXR activity in the body and alters the fecal BA profile, leading to reduced internal lipid absorption and reduced hyperglycemia [93-95]. Insufficient expression of FXR in intestine of mice is resistant to insulin resistance, NAFLD and obesity induced by HFD, which confirms the possibility that intestinal FXR is associated with the development of metabolic diseases. HFD-fed mice were given Glycine-β-muricholic acid (Gly-βMCA) a potent intestinal FXR antagonist that is resistant to BSH, to simulate the effects of changes in the mimic microbiota on HFD-induced metabolic diseases. The study have shown that Gly-β-MCA reduces the poor metabolic phenotype due to lower serum ceramide. Intestinal FXR deficiency in mice also leads to a decrease in serum ceramide, a adaptation to metabolism and resistance to HFD-induced metabolic diseases, which can be reversed by injection of C16:0 ceramide. Due to the role of endogenous hepatic FXR agonists, its target genes related to ceramide synthesis in the ileum of mice are activated, which are inhibited by a decrease in serum ceramide when Gly-β-MCA is controlled. It shows that Ceramide produced in the ileum under FXR management is associated with metabolic diseases [96]. Jiang has demonstrated that it is possible to intervene in the development of NAFLD associated with the gut microbiota by inhibiting the intestinal FXR-ceramide axis, which correlates the microbiome, nuclear receptor signaling and NAFLD [97]. Consequently, Gly-β-MCA can inhibit FXR receptors in the intestine and prevent and treat fatty liver [98].
Lili Sheng et al. had showed that BA and FXR and related intestinal microbiota are concerned with differences of gender in metabolic diseases [99]. Gender differences in predicted microbiota function, insulin sensitivity, and steatosis induced by WD were attributed to FXR. The lack of FXR enriched Helicobacteraceae, Deferribacteraceae and Desulfovibrionaceae, which were accompanied by increased and serum lipids as well as β-muricholic acid and taurine-conjugated cholic acid in the liver. The FXR signal in the ileum can also modulate the composition of bile acid via enterobacterial-mediated degradation. Enterobacteriaceae enhance ileal FXR signaling by degrading (deconjugated) taurocholic acid (TCA) [100]. However, the specific role of evaluating individual bile acid is hampered by the fact that the conversion of bile acid occurs under the action of Enterobacteriaceae-mediated degradation, dehydroxylation, and amino acid binding in the liver and the rehydroxylation pathway [101]. Fexaramine, an enteric-restricted FXR agonist that restores FXR activity in the small intestine of chronically fed mice, which protects mice from liver damage induced by alcohol. Although the metabolism of bile acids is slightly altered, fexaramine treatment stabilizes the intestinal barrier and significantly regulates liver genes that affect lipid metabolism. In conclusion, alcohol-related metagenomic alterations result in changes in the bile acid profile. Effective interventions modulate bile acid-FXR-FGF15 signaling and protected mice from liver alcohol-induced disease by regulating hepatic Cyp7al and lipid metabolism [102].
TGR5 that is expressed in a variety of tissues including intestine, gallbladder, liver, and brain is related to cAMP and determined to affect energy homeostasis, immune response, insulin secretion, inflammation and gallbladder relaxation [103-110]. In recent years, TGR5 has become a promising intervention target [111,112]. In the intestine, Tgr5 is activated by its specific agonist to protect intestinal barrier function and from inflammation, stimulate the gallbladder to refill, stimulate glucagon, such as peptide 1 (GLP-1) secreted from the intestinal secretion of cells, thereby stimulating insulin secretion from pancreatic cells [113,114]. In brown adipocytes, activated TGR5 stimulates thyroid hormones by inducing thyroid hormone deiodinase 2 (DIO2) to promote energy metabolism [115]. Secondary bile acids are potent activators of TGR5, such as DCA and LCA, which induce cAMP and activate protein kinase alpha signaling pathways [103,104]. Activation of TGR5 can maintain glucose homeostasis to attenuate hepatic steatosis and obesity in obese mice induced by diet [116]. Bile acid inhibited NLRP3 inflammasome activation via the TGR5-cAMP-PKA axis. Furthermore, studies in vivo have shown that bile acids and TGR5 activation blocked NLRP3 inflammasome-dependent inflammation, such as type-2 diabetes-related inflammation, lipopolysaccharide-induced systemic inflammation [117]. Activation of intestinal in the intestine promotes elevated levels of LCA in the gut to affect bile acid metabolism. Simultaneously, the increased LCA activate TGR5 in the gut, follow which facilitates GLP-1 secretion from intestinal L cells to lead adipose tissue browning and regulate lipid and glucose metabolism in the liver [118]. The gastrointestinal FXR-gut microbiota-TGR5-GLP-1 axis is a key target for regulating intestinal bile acid receptor signaling to improve homeostasis and liver metabolism. The fecal and mucosa-associated microbiota in animals and humans with alcoholic liver disease is also been altered [119-122]. Alcoholic fatty liver has been shown to be connected with elevated levels of bacterial choloylglycine hydrolase, which deconjugates intestine bile acids in mice by analyzing the gut microbiota and bile acid. Overexpression of the CYP7A1 protein reduces FXR activity in intestinal epithelial cells, which inhibits FGF-15 protein secretion. The total concentration of bile acids in the plasma is elevated, which is likely to cause hepatocyte death together with ethanol. Furthermore, consumption of symbiotic microbiota with non-absorbable antibiotics reduced the expression of Cyp7a1 gene in liver to reduce the incidence of alcoholic liver disease in mice, which suggested that the increase in bile acid synthesis is dependent on gut microbiota [123].
Taken together, the gut microbiota can participate regulation and metabolism of bile acids composition and metabolism, and affect the occurrence and development of diseases through regulation. In addition, the intestinal microflora can effectively hydrolyze the bound parasites or heterologous organisms that have been cleared by bile acid. Substances and promote activation of these substances or enterohepatic circulation .In addition, the intestinal microbial flora can also be very effective in the hydrolysis of bound parasites or heterologous substances in organisms that have been cleared by bile acid and promote the activation of these substances or enterohepatic circulation [124].
Effect of bile acid on intestinal flora
The relationship between bile acids and intestinal flora is not unidirectional. Bile acids can alter the structure of the intestinal flora by mediating bacterial growth and inhibiting the growth of other sensitive bacteria [90,91,125]. Taurine bile salts, a bile salt hydrolysate,can inhibit a certain dose of C. difficile (C. difficile) toxins, thereby effectively avoiding damage to colonic epithelial cells [126]. Intestinal purification can stabilize the intestinal barrier, prevent excessive growth of intestinal bacteria and bacterial transfer to the liver [127]. Bile acid act as a detersive agent, which can destroy the bacteria’s cell membrane and exert a bacteriostatic effect directly, and can indirectly affect the gut microbiota through FXR [91]. FXR knockout mice exhibit higher bacterial density and impaired epithelial barrier integrity in the ileum [128]. HFD-induced mice knocked out of FXR, rather than wild-type control mice, increased the amount of Phylum Firmicutes and reduced Bacteroides levels [129]. Increased hydrophobicity of bile acid can lead to increased antigen and bacterial translocation in the intestine, resulting in the development of diseases such as cholelithiasis, inflammatory bowel disease, and colon cancer [130]. When feeding bile acid, complex and significant changes were observed in the intestine. In general, bile acid affect the gut microbiota diversity and regulate the structure of gut microbiota. Islam et al. had demonstrated that the intake of moderate amounts of bile acid (0.00125 mol/L) and high bile acid (0.005 mol/L) caused changes in the gut microbiota, in particular the significant expansion of the genus Mycobacteria [131]. The response of the gut microbiota to cholic acid differed from that of feeding for 50 days, possibly due to differences in dose and exposure time. In addition, there seems to be a difference in the response of gut microbes to bile acid. Metabolic pathway associated with bile acids is one of the most relevant pathways involved in antibiotic-induced dysregulation of the gut microbiota. Cholic acid, deoxycholic acid, and chenodeoxycholic acid were observed in the fecal samples of rats with dysbacteriosis in the intestine. After CSGS treatment, all derivatives of the bile acid metabolites were corrected, explaining that bile acid metabolism involves dysregulation of the gut microbiota [132]. In another study, colesevelam promotes bile acid excretion through feces and conversion to secondary bile acids, while liver and biliary BA components change to hydrophilic BA.The expression of ileal FGF15 is eliminated by colesevelam to stimulate the secretion of GLP-1 from enteroendocrine l-cells and attenuates liver and bile duct injury in Mdr2-/- mice. Microbiota analysis showed that the level of the phylum δ-Proteobacteria was increased and Clostridiales was transformed into Lactobacillus within the phyla Firmicutes after colesevelam treatment [133]. In addition, bile acid has been shown to mediate host resistance to Clostridium infection. The reduction of primary and secondary bile acid in the intestinal tract after morphine treatment is associated with an increase in pathogenic intestinal bacteria such as Enterococcus faecalis. Bile acid was negatively correlated with enterococci and erysipelas, but positively correlated with Bacteroideae [134].
Conclusion and future development
Bile acids can act as important signaling molecules through different cell signaling pathways and nuclear receptor to regulate glucose, lipid and energy metabolism. Bile acid disorders can lead to a variety of disease states, including metabolic diseases such as diabetes, gastrointestinal diseases, liver diseases, and inflammation and cancer. This review highlights the signaling and regulation involved in bile acid synthesis and metabolic pathways. In addition, the interaction between bile acids and the gut microbiota is also discussed.
Recently, metagenomics and bioinformatics analysis have shown that the intestinal microbiota can regulate the synthesis and metabolism of bile acids, and also reveals that the regulation of gut microbiota can prevent or treat metabolic diseases associated with bile acids. Bile acids and their activated nuclear receptors (such as FXR) and membrane receptors (TGR5) are key therapeutic targets for related diseases. Regulating the gut microbiota to activate the tgr5-glp-1 signal is a new direction in the treatment of diseases such as NAFLD, diabetes and obesity.
In addition, individualized studies of gut microbiota also contribute to disease treatment. A study has found that the treatment of type 2 diabetes with acarbose depends on the composition of the gut microbiota before treatment. Compared with the predominant gut microbiota of Prevotella, those strains with high abundance Bacteroides had greater changes in plasma BA after acarbose treatment and improved metabolic parameters. Thus, the patient can be stratified based on the composition of the gut microbiota before treatment to facilitate a more rational customized treatment [99]. Another study suggests that probiotics or other means of preventing and treating metabolic disorders should be adjusted based on patient gender and other phenotypes (eg, microbiota and BA profiles) [99]. However, artificial disturbance of the gut microbiota or bile acid composition may cause side effects, such as affecting the enterohepatic circulation of bile acids or breaking the homeostasis in the organism, which needs further study.
Bacteria in humans and animals are subjected to environmental stress such as stomach acid and intestinal bile acids.Adaptation and tolerance to bile acid toxicity are key conditions for the survival of microbiota in the gut of humans and animals. Some bile acids, such as LCA, DCA, CDCA and TCNCA, may promote cancer or inhibit cancer because of their cytotoxicity, and the mechanism of action needs further discussion [52,132-136]. Prevention of bile acid toxicity by regulating the amount and type of bile acids in the liver and intestine will require a better understanding of the pathway of bile acid synthesis, transport and metabolism.
With the advent of new analytical tools and strategies such as metabolomics and metagenomics, the interaction between bile acids and gut microbiota and its regulation of disease-related signaling pathways are rapidly gaining insight. The regulation of bile acid profiles and gut microbiota presents new treatments for the treatment of metabolic and digestive tract diseases. There are still three core challenges to establish the relationship between gut microbiota-metabolite-host health and linkages at the molecular level. Firstly, analyze the biological activities of these metabolites, including their association with human diseases; secondly, identify the metabolic pathways that produce these metabolites; thirdly, use the analytical results to further combine genetic engineering and synthetic biology to reprogram these Metabolite produced by intestinal microbes. This strategy has great promise and clearly indicates the next direction in the study of these diseases. Due to the complexity of the bile acid signaling pathway and the complex metabolic interactions between the host and the gut microbiota, future research should understand bile acid-microbiota crosstalk and its effects on related diseases based on systems biology. Future research to prevent and treat disease may include identifying beneficial symbiotic microbiota and its key metabolites that drive the development of related diseases, especially inflammation and cancer.
This work was supported by grants from the Key Program of Natural Science Foundation of State (Grant No. 81830110, 81430093, 81373930, 81673586, 81703685, 81302905, 81503386), National Key Subject of Drug Innovation (Grant No. 2015ZX09101043-005, 2015ZX09101043-011), TCM State Administration Subject of Public Welfare of (Grant No. 2015468004), Major Projects of Application Technology Research and Development Plan in Heilongjiang Province (GX16C003), TCM State Administration Subject of Public Welfare (Grant No. 2015468004), Specialized Research Fund for the Doctoral Program of Higher Education (20132327130001, 20122327120006), University Nursing Program for Young Scholars with Creative Talents in Heilongjiang Province (UNPYSCT-2015118, UNPYSCT-2016213), Young Talent Lift Engineering Project of China Association of Traditional Chinese Medicine (QNRC2-B06), Outstanding Talents Foundation of Heilongjiang University of Chinese Medicine (2018jc01), Doctoral Innovation Fund of Heilongjiang University of Chinese Medicine (2018bs02), Foundation of Heilongjiang University of Chinese Medicine (Grant no. 201209), University Nursing Program for Young Scholars with Creative Talents in Heilongjiang Province (UNPYSCT-2015118, UNPYSCT-2016213, UNPYSCT-2016212), Application Technology and Development of Youth Talents Project in Harbin (2014RFQXJ116), Chinese Postdoctoral Science Foundation (2017M621319b), Returned Oversea Scholars Program of Heilongjiang Province (2017QD0025), Natural Science Foundation of Heilongjiang Province of Chi na (QC2018117).
- Dodd D, Spitzer MH, Treuren WV, Merrill BD, Hryckowian AJ, et al. (2017) A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551: 648-652. Link: https://goo.gl/qUVYQu
- Hall AB, Tolonen AC, Xavier RJ (2017) Human genetic variation and the gut microbiome in disease. Nature Reviews Genetics 18: 690-699. Link: https://goo.gl/HgJng6
- Wang X, Zhang A, Wang P (2013) Metabolomics Coupled with Proteomics Advancing Drug Discovery toward More Agile Development of Targeted Combination Therapies. Molecular Cellular Proteomics 12: 1226-1238. Link: https://goo.gl/eTBXue
- Zhang A, Sun H, Wang X (2014) Potentiating therapeutic effects by enhancing synergism based on active constituents from traditional medicine. Phytother Res 28: 526-533. Link: https://goo.gl/4QDpBF
- Guo C, Xie S, Chi Z, Zhang J, Liu Y, et al. (2016) Bile acids control inflammation and metabolic disorder through inhibition of nlrp3 inflammasome. Science Foundation in China 45: 802-816. Link: https://goo.gl/FL7NYE
- Hao H, Cao L, Jiang C, Che Y, Zhang S, et al. (2017) Farnesoid x receptor regulation of the nlrp3 inflammasome underlies cholestasis-associated sepsis. Cell Metabolism 25: 856-867. Link: https://goo.gl/RXbPgC
- Zarrinpar A, Chaix A, Xu ZZ, Chang MW, Marotz CA, et al. (2018) Antibiotic-induced microbiome depletion alters metabolic homeostasis by affecting gut signaling and colonic metabolism. Nature Communications 9: 2872. Link: https://goo.gl/RMS9rf
- Chiang JY (2013) Bile acid metabolism and signaling. Comprehensive Physiology 3: 1191-1212. Link: https://goo.gl/dC6DBB
- Copple BL, Li T (2016) Pharmacology of bile acid receptors: evolution of bile acids from simple detergents to complex signaling molecules. Pharmacological Research 104: 9-21. Link: https://goo.gl/vQfmgE
- Donepudi AC, Boehme S, Li F, Chiang JY (2016) G protein-coupled bile acid receptor plays a key role in bile acid metabolism and fasting-induced hepatic steatosis. Hepatology 65: 813-827. Link: https://goo.gl/ydr6uA
- Desai MS, Mathur B, Eblimit Z, Vasquez H, Taegtmeyer H, et al. (2016) Bile acid excess induces cardiomyopathy and metabolic dysfunctions in the heart. Hepatology 65: 189-201. Link: https://goo.gl/1P3eDq
- XN Li, Zhang A, Wang M, Sun H, Liu Z, et al. (2017) Screening the active compounds ofPhellodendriAmurensis cortex for treating prostate cancer by high-throughput chinmedomics. Sci Rep 7: 46234. Link: https://goo.gl/dAiVDv
- Zhang K, Yan G, Zhang A, Sun H, Wang X (2017) Recent advances in pharmacokinetics approach for herbal medicine. RSC Advances 7: 28876-28888. Link: https://goo.gl/yiyifq
- Sun H, Wu F, Zhang A, Wei W, Han Y, et al. (2013) Profiling and identification of the absorbed constituents and metabolites of Schisandra lignans by ultra-performance liquid chromatography coupled to mass spectrometry. Biomed Chromatogr 27: 1511-1519. Link: https://goo.gl/qPw6SQ
- Li X, Sun H, Zhang A (2017) High-throughput LC-MS method for the rapid characterization of multiple chemical constituents and metabolites of Da-Bu-Yin-Wan. J Sep Sci 40: 4102-4112. Link: https://goo.gl/o7b5xB
- Ma C, Han M, Heinrich B, Fu Q, Zhang Q, et al. (2018) Gut microbiome-mediated bile acid metabolism regulates liver cancer via nkt cells. Science 360: eaan5931. Link: https://goo.gl/sFF5X6
- Cao W, Kayama H, Chen ML, Delmas A, Sun A, et al. (2017) The xenobiotic transporter mdr1 enforces t cell homeostasis in the presence of intestinal bile acids. Immunity 47: 1182-1196. Link: https://goo.gl/y9sZ2F
- Catry E, Bindels LB, Tailleux A, Lestavel S, Neyrinck AM, et al. (2017) Targeting the gut microbiota with inulin-type fructans: preclinical demonstration of a novel approach in the management of endothelial dysfunction. Gut 67: 271-283. Link: https://goo.gl/7Hr7vh
- Zhang A, Sun H, Qiu S (2013) Advancing drug discovery and development from active constituents of yinchenhao tang, a famous traditional chinese medicine formula. Evidence-Based Complementray and Alternative Medicine 2013: 257909. Link: https://goo.gl/TwnwWS
- Chiang JYL (2009) Bile acids: regulation of synthesis. Journal of Lipid Research 50: 1955-1966. Link: https://goo.gl/kgDWgp
- Zhang A, Sun H, Wang X (2012) Serum metabolomics as a novel diagnostic approach for disease: A systematic review. Anal Bioanal Chem 404: 1239-1245. Link: https://goo.gl/E3kUP6
- Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B (2009) Role of bile acids and bile acid receptors in metabolic regulation. Physiological Reviews 89: 147-191. Link: https://goo.gl/9Akm8h
- Sun H, Liu J, Zhang A (2016) Characterization of the multiple components of AcanthopanaxSenticosus stem by ultra-high performance liquid chromatography with quadrupole time-of-flight tandem mass spectrometry. Journal of Separation Science 39: 496-502. Link: https://goo.gl/gT9gbq
- Gonzalez FJ (2012) Nuclear receptor control of enterohepatic circulation. Comprehensive Physiology 2: 2811-2828. Link: https://goo.gl/uauewc
- Zhang Y, Zhang A, Zhang Y, Sun H, Meng X, et al. (2016) Application of Ultra-performance Liquid Chromatography with Time-of-Flight Mass Spectrometry for the Rapid Analysis of Constituents and Metabolites from the Extracts of Acanthopanaxsenticosus Harms Leaf. Pharmacogn Mag 12: 145-152. Link: https://goo.gl/ajSn5d
- Zhang A, Sun H, Yan G (2017) Recent developments and emerging trends of mass spectrometry for herbal ingredients analysis. Trac Trends in Analytical Chemistry 94: 70-76. Link: https://goo.gl/WPCSwq
- Wang X, Sun H, Zhang A, Wang P, Han Y (2011) Ultra-performance liquid chromatography coupled to mass spectrometry as a sensitive and powerful technology for metabolomic studies. Sep Sci 34: 3451-3459. Link: https://goo.gl/wEAqFJ
- Li T, Chiang JY (2014) Bile acid signaling in metabolic disease and drug therapy. Pharmacological Reviews 66: 948-983. Link: https://goo.gl/ZTQmPi
- Zhang A, Sun H, Wang X (2014) Urinary metabolic profiling of rat models revealed protective function of scoparone against alcohol induced hepatotoxicity. Sci Rep 4: 6768. Link: https://goo.gl/VSXkPJ
- Wahlström A, Sayin SI, Marschall HU, Bäckhed F (2016) Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metabolism 24: 41-50. Link: https://goo.gl/Nrx7TE
- Wang X, Lv H, Zhang A (2014) Metabolite profiling and pathway analysis of acute hepatitis rats by UPLC-ESI MS combined with pattern recognition methods. Liver Int 34: 759-770. Link: https://goo.gl/uBPFh4
- Kliewer SA, Mangelsdorf DJ (2015) Bile acids as hormones: the fxr-fgf15/19 pathway. Digestive Diseases 33: 327-331. Link: https://goo.gl/mQ7aQe
- Qiu S, Zhang AH, Sun H, Yan GL, Wang XJ (2014) Overview on metabolomics in traditional Chinese medicine. World J Pharmacol 3: 33-38. Link: https://goo.gl/zyCo5U
- Wang X, Zhang A, Yan G (2014) UHPLC-MS for the analytical characterization of traditional Chinese medicines. Trac Trends in Analytical Chemistry 63: 180-187. Link: https://goo.gl/cVTZ37
- Zhang A, Sun H, Wang X (2018) Mass spectrometry-driven drug discovery for development of herbal medicine. Mass Spectrom Rev 37: 307-320. Link: https://goo.gl/mCK4vd
- Dawson PA (2011) Role of the intestinal bile acid transporters in bile acid and drug disposition. Handbook of Experimental Pharmacology 201: 169-203. Link: https://goo.gl/Rk5Hcv
- Xue C, Zhang A, Sun H (2014) An improved ultra-performance liquid chromatography-electrospray ionization/quadrupole-time-of-flight high-definition mass spectrometry method for determining ingredients of herbal Fructus corni in blood samples. Pharmacognosy Magazine 10: 422-429. Link: https://goo.gl/HsHMer
- Li XN, Zhang A, Sun H, Song Y, Zou D, et al. (2016) Rapid discovery of absorbed constituents and metabolites in rat plasma after the oral administration of Zi Shen Wan using high-throughput UHPLC-MS with a multivariate analysis approach.Sep Sci 39: 4700-4711. Link: https://goo.gl/2tEbSR
- Jahn D, Rau M, Hermanns HM, Geier A (2015) Mechanisms of enterohepatic fibroblast growth factor 15/19 signaling in health and disease. Cytokine & Growth Factor Reviews 26: 625-635. Link: https://goo.gl/PPNXdD
- Zhang AH, Yu JB, Sun H, Kong L, Wang XQ, et al. (2018) Identifying quality-markers from Shengmai San protects against transgenic mouse model of Alzheimer's disease using chinmedomics approach. Phytomedicine. 45: 84-92. Link: https://goo.gl/u3TMpo
- Zhou X, Cao L, Jiang C, Xie Y, Cheng X, et al. (2014) Pparα-ugt axis activation represses intestinal fxr-fgf15 feedback signalling and exacerbates experimental colitis. Nature Communications 5: 4573. Link: https://goo.gl/7wssBt
- Sun H, Zhang A, Yang L, Li M, Fang H, et al. (2018) High-throughput chinmedomics strategy for discovering the quality-markers and potential targets for Yinchenhao decoction. Phytomedicine. pii: S0944-7113(18)30115-6. Link: https://goo.gl/1osTcx
- Wang X, Zhang A, Kong L, Yu J, Gao H, et al. (2017) Rapid discovery of quality-markers from Kaixin San using chinmedomics analysis approach. Phytomedicine. pii: S0944-7113(17)30188-5. Link: https://goo.gl/rKLMYg
- Pathak P, Liu H, Boehme S, Xie C, Krausz KW, et al. (2017) Farnesoid x receptor induces takeda g-protein receptor 5 crosstalk to regulate bile acid synthesis and hepatic metabolism. Journal of Biological Chemistry 292: 11055-11069. Link: https://goo.gl/kRYzUK
- Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, et al. (2013) Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. Gut Microbes 58: 949-955. Link: https://goo.gl/73Fg7S
- Kuipers F, Groen AK (2017) An unexpected role for bile acid synthesis in adaptation to low temperature. Nature Medicine 23: 800-802. Link: https://goo.gl/zEZ4av
- Wang X, Zhang A, Zhou X (2016) An integrated chinmedomics strategy for discovery of effective constituents from traditional herbal medicine. Scientific Reports 6. Link: https://goo.gl/sAumvn
- Fuchs CD, Paumgartner G, Mlitz V, Kunczer V, Halilbasic E, et al. (2018) Colesevelam attenuates cholestatic liver and bile duct injury in mdr2 -/-, mice by modulating composition, signalling and excretion of faecal bile acids. Gut 67:1683-1691. Link: https://goo.gl/6q6Jmv
- Zhang A, Liu Q, Zhao H (2016) Phenotypic characterization of nanshi oral liquid alters metabolic signatures during disease prevention. Scientific Reports 6. Link: https://goo.gl/J9PCJc
- Pathak P, Cen X, Nichols RG, Ferrell JM, Boehme S, et al. (2018) Intestine farnesoid x receptor agonist and the gut microbiota activate g‐protein bile acid receptor‐1 signaling to improve metabolism. Hepatology 68: 1574-1588. Link: https://goo.gl/e3SqSd
- Han Y, Zhang A, Sun H, Zhang Y, Meng X, et al. (2017) High-throughput ultra-high performance liquid chromatography coupled to quadrupole time-of-flight mass spectrometry method or the rapid analysis and characterization of multiplenstituents of Radix Polygalae.Sep.Sci 40: 2178-2187. Link: https://goo.gl/LRgxJD
- Ma C, Han M, Heinrich B, Fu Q, Zhang Q, et al. (2018) Gut microbiome–mediated bile acid metabolism regulates liver cancer via nkt cells. Science 360. pii: eaan5931. Link: https://goo.gl/TdSUWe
- Chu H, Zhang A, Han Y, Lu S, Kong L, et al. (2016) Metabolomics approach to explore the effects of Kai-Xin-San on Alzheimer's disease using UPLC/ESI-Q-TOF mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 1015-1016: 50-61. Link: https://goo.gl/1XVnEe
- Swann JR, Want EJ, Geier FM, Spagou K, Wilson ID, et al. (2011) Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc Natl Acad Sci U S A 108: 4523-4530. Link: https://goo.gl/NF4SHn
- Qiu S, Zhang A, Zhang T, Sun H, Guan Y, et al. (2017) Dissect new mechanistic insights for geniposide efficacy on the hepatoprotection using multiomics approach. Oncotarget 8: 108760-108770. Link: https://goo.gl/oeYsvg
- Hartmann P, Hochrath K, Horvath A, Chen P, Seebauer CT, et al. (2018) Modulation of the intestinal bile acid-fxr-fgf15 axis improves alcoholic liver disease in mice. Hepatology 67: 2150-2166. Link: https://goo.gl/31LMvq
- Zhang A, Zhou X, Zhao H, Zou S, Ma CW, et al. (2017) Metabolomics and proteomics technologies to explore the herbal preparation affecting metabolic disorders using high resolution mass spectrometry. Mol Biosyst 13: 320-329. Link: https://goo.gl/mECaod
- Jiang JK, Hang XM, Min Z, Liu XL, Li DT, et al. (2010) Diversity of bile salt hydrolase activities in different lactobacilli toward human bile salts. Annals of Microbiology 60: 81-88. Link: https://goo.gl/RxwwHP
- Jiang C, Xie C, Li F, Zhang L, Nichols RG, et al. (2015) Intestinal farnesoid x receptor signaling promotes nonalcoholic fatty liver disease. Journal of Clinical Investigation 125: 386-402. Link: https://goo.gl/mofwcX
- Yang JY, Lee YS, Kim Y, Lee SH, Ryu S, et al. (2016) Gut commensal bacteroides acidifaciens prevents obesity and improves insulin sensitivity in mice. Mucosal Immunology 10: 104-116. Link: https://goo.gl/mhinJb
- Zhang A, Sun H, Wang X (2017) Emerging role and recent applications of metabolomics biomarkers in obesity disease research. RSC Advances 7: 14966-14973. Link: https://goo.gl/PLF6Yb
- Theriot CM, Koenigsknecht MJ, Jr CP, Hatton GE, Nelson AM, et al. (2014) Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to clostridium difficile infection. Nature Communications 5: 3114. Link: https://goo.gl/dcR1Xs
- Zhang A, Fang H, Wang Y, Yan G, Sun H, et al. (2017) Discovery and verification of the potential targets from bioactive molecules by network pharmacology-based target prediction combined with high-throughput metabolomics. RSC Adv 7: 51069-51078. Link: https://goo.gl/J2sWEM
- Yin Q, Wang P, Zhang A, Sun H, Wu X, et al. (2013) Ultra-performance LC-ESI/quadrupole-TOF MS for rapid analysis of chemical constituents of Shaoyao-Gancao decoction. J Sep Sci 36: 1238-1246. Link: https://goo.gl/7Zbux5
- Del CF, Nobili V, Vernocchi P, Russo A, Stefanis C, et al. (2017) Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology 65: 451-464. Link: https://goo.gl/6i69jw
- Dong W, Wang P, Meng X, Sun H, Zhang A, et al. (2012) Ultra-performance liquid chromatography-high-definition mass spectrometry analysis of constituents in the root of Radix Stemonae and those absorbed in blood after oral administration of the extract of the crude drug. Phytochem Anal 23: 657-667. Link: https://goo.gl/dzT2yf
- Shapiro H, Suez J, Elinav E (2017) Personalized microbiome-based approaches to metabolic syndrome management and prevention. Journal of Diabetes 9: 226-236. Link: https://goo.gl/Aa3bv8
- Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson, et al. (2009) Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci 106: 14728-14733. Link: https://goo.gl/uNixhz
- Han Y, Zhang A, Sun H (2017) High-throughput ultra-high performance liquid chromatography combined with mass spectrometry approach for the rapid analysis and characterization of multiple constituents of the fruit of Acanthopanaxsenticosus (Rupr. et Maxim.) Harms. Journal of Separation Science 40: 2178-2187. Link: https://goo.gl/LRhEPq
- Marika Z, Petia KD, Markiewicz LH, Marcus S, Kozak LP, et al. (2016) Altered microbiota contributes to reduced diet-induced obesity upon cold exposure:. Cell Metabolism 23: 1216-1223. Link: https://goo.gl/j2Y1mz
- Worthmann A, John C, Rühlemann MC, Baguhl M, Heinsen FA, et al. (2017) Cold-induced conversion of cholesterol to bile acids in mice shapes the gut microbiome and promotes adaptive thermogenesis. Nature Medicine 23: 839-849. Link: https://goo.gl/D1x5Je
- Liu JH, Sun H, Zhang AH (2014) Serum pharmacochemistry combined with multiple data processing approach to screen the bioactive components and their metabolites in Mutan Cortex by ultra-performance liquid chromatography tandem mass spectrometry. Biomedical Chromatography 28: 500-510. Link: https://goo.gl/Lnfx27
- Buffie CG, Bucci V, Stein RR, Mckenney PT, Ling L, et al. (2015) Precision microbiome reconstitution restores bile acid mediated resistance to clostridium difficile. Nature 517: 205-208. Link: https://goo.gl/o2hUwU
- Sun H, Yin Q, Zhang A, Wang X (2012) UPLC–MS/MS performing pharmacokinetic and biodistribution studies of rhein. Journal of separation science 35: 2063-2068. Link: https://goo.gl/qq3Bo2
- Sun L, Xie C, Wang G, Wu Y, Wu Q, et al. (2018) Gut microbiota and intestinal fxr mediate the clinical benefits of metformin. Nature Medicine 24: 1919-1929. Link: https://goo.gl/UWhnq4
- Bernstein H, Bernstein C, Payne CM, Dvorak K (2009) Bile acids as endogenous etiologic agents in gastrointestinal cancer. World Journal of Gastroenterology 15: 3329-2240. Link: https://goo.gl/TNe3DL
- Han Y, Wu F, Zhang A (2015) Characterization of multiple constituents in rat plasma after oral administration of Shengmai San using ultra-performance liquid chromatography coupled with electrospray ionization/quadrupole-time-of-flight high-definition mass spectrometry. Analytical Methods 7: 830-837. Link: https://goo.gl/zRtUkA
- Bernstein C, Holubec H, Bhattacharyya AK, Nguyen H, Payne CM, et al. (2011) Carcinogenicity of deoxycholate, a secondary bile acid. Archives of Toxicology 85: 863-871. Link: https://goo.gl/p2zvHn
- Xie G, Wang X, Huang F, Zhao A, Chen W, et al. (2016) Dysregulated hepatic bile acids collaboratively promote liver carcinogenesis. International Journal of Cancer, 139(8), 1764-1775.68. Xie, G. et al. Distinctly altered gut microbiota in the progression of liver disease. Oncotarget 7: 19355–19366. Link: https://goo.gl/KfH9Qt
- Yang B, Dong W, Zhang A, Sun H, Wu F, et al. (2011) Ultra‐performance liquid chromatography coupled with electrospray ionization/quadrupole‐time‐of‐flight mass spectrometry for rapid analysis of constituents of Suanzaoren decoction. Journal of separation science 34: 3208-3215. Link: https://goo.gl/L9txSB
- Venkatesh, Madhukumar, Mukherjee, Subhajit, Wang, & Hongwei, et al. (2014) Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor pxr and toll-like receptor 4. Immunity 41: 296-310. Link: https://goo.gl/izfzY3
- Sun H, Wu F, Zhang A, Wei W, Han Y, et al. (2013) Pharmacokinetic study of schisandrin, schisandrol B, schisantherin A, deoxyschisandrin, and schisandrin B in rat plasma after oral administration of S hengmaisan formula by UPLC‐MS. Journal of separation science 36: 485-491. Link: https://goo.gl/TJydLf
- Swidsinski A, Loeningbaucke V, Vaneechoutte M, Doerffel Y (2010) Active crohn's disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflammatory Bowel Diseases 14: 147-161. Link: https://goo.gl/FvVkgy
- Sun H, Dong T, Zhang A, Yang J, Yan G, et al. (2013) Pharmacokinetics of hesperetin and naringenin in the Zhi Zhu Wan, a traditional Chinese medicinal formulae, and its pharmacodynamics study. Phytotherapy Research 27: 1345-1351. Link: https://goo.gl/u86tY1
- Vassileva G, Golovko A, Markowitz L, Abbondanzo SJ, Zeng M, et al. (2006) Targeted deletion of gpbar1 protects mice from cholesterol gallstone formation. Biochemical Journal: 398: 423-430. Link: https://goo.gl/Lw2AQ9
- Zhao Q, Zhang A, Zong W (2017) Exploring potential biomarkers and determining the metabolic mechanism of type 2 diabetes mellitus using liquid chromatography coupled to high-resolution mass spectrometry. Rsc Advances 7: 44186-44198. Link: https://goo.gl/KpgRxu
- Devkota S, Wang Y, Musch MW, Leone V, Fehlnerpeach H, et al. (2012) Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in il10-/- mice. Nature 142: 104-108. Link: https://goo.gl/PFEfwT
- Dawson PA, Karpen SJ (2015) Intestinal transport and metabolism of bile acids. Journal of Lipid Research 56: 1085-1099. Link: https://goo.gl/PRRASi
- Degirolamo C, Rainaldi S, Bovenga F, Murzilli S, Moschetta A (2014) Microbiota modification with probiotics induces hepatic bile acid synthesis via downregulation of the fxr-fgf15 axis in mice. Cell Reports 7: 12-18. Link: https://goo.gl/iuiU29
- Sun H, Zhang Hl (2018) Network pharmacology combined with functional metabolomics discover bile acid metabolism as a promising target for mirabilite against colorectal cancer. RSC Adv 8: 30061-30070. Link: https://goo.gl/JYCBr7
- Parséus A, Sommer N, Sommer F, Caesar R, Molinaro A, et al. (2017) Microbiota-induced obesity requires farnesoid x receptor. Gut 66: 429-437. Link: https://goo.gl/i7KBHe
- Wang X, Zhang A, Yan G, Sun W, Han Y, et al. (2013) Metabolomics and proteomics annotate therapeutic properties of geniposide: targeting and regulating multiple perturbed pathways. PLoS One 8: e71403. Link: https://goo.gl/ehv6Pu
- Wang X, Zhang A, Hui S (2016) Discovery and development of innovative drug from traditional medicine by integrated chinmedomics strategies in the post-genomic era. Trends in Analytical Chemistry 76: 86-94. Link: https://goo.gl/65T4qK
- Zhang Y, Lee FY, Barrera G, Lee H, Vales C, et al. (2006) Activation of the nuclear receptor fxr improves hyperglycemia and hyperlipidemia in diabetic mice. Proceedings of the National Academy of Sciences of the United States of America 103: 1006-1011. Link: https://goo.gl/yDngQw
- Zhang A, Zou D, Yan G, Tan Y, Sun H, et al. (2014) Identification and characterization of the chemical constituents of Simiao Wan by ultra high performance liquid chromatography with mass spectrometry coupled to an automated multiple data processing method. Journal of separation science 37: 1742-1747. Link: https://goo.gl/Y8xXi4
- Gonzalez FJ, Jiang C, Patterson AD (2016) An intestinal microbiota-farnesoid x receptor axis modulates metabolic disease. Gastroenterology 151: 845-859. Link: https://goo.gl/7c7ZgC
- Zhang T, Zhang A, Qiu S (2016) High-throughput metabolomics approach reveals new mechanistic insights for drug response of phenotypes of geniposide towards alcohol-induced liver injury by using liquid chromatography coupled to high resolution mass spectrometry. Molecular Biosystems 73-82. Link: https://goo.gl/6oYfva
- Jiang C, Xie C, Lv Y, Li J, Krausz KW, et al. (2015) Intestine-selective farnesoid x receptor inhibition improves obesity-related metabolic dysfunction. Nature Communications 6: 10166. Link: https://goo.gl/mf8EjA
- Sheng L, Jena PK, Liu HX, Kalanetra KM, Gonzalez FJ, et al. (2017) Gender differences in bile acids and microbiota in relationship with gender dissimilarity in steatosis induced by diet and fxr inactivation. Scientific Reports 7: 1748. Link: https://goo.gl/V1i6R2
- Zhang T, Zhang A, Qiu S (2016) Current Trends and Innovations in Bioanalytical Techniques of Metabolomics. Critical Reviews in Analytical Chemistry 46: 342-351. Link: https://goo.gl/5CdDM8
- Zhang Y, Klaassen CD (2010) Effects of feeding bile acids and a bile acid sequestrant on hepatic bile acid composition in mice. Journal of Lipid Research 51: 3230-3242. Link: https://goo.gl/22vmAu
- Hartmann P, Hochrath K, Horvath A, Chen P, Seebauer CT, et al. (2017) Modulation of the intestinal bile acid-fxr-fgf15 axis improves alcoholic liver disease in mice. Hepatology.
- Liu Q, Zhang A, Wang L (2016) High-throughput chinmedomics-based prediction of effective components and targets from herbal medicine AS1350. Scientific Reports 6: 38437. Link: https://goo.gl/25xos9
- Maruyama T, Miyamoto Y, Nakamura T, Tamai Y, Okada H, et al. (2002) Identification of membrane-type receptor for bile acids (m-bar). Biochem Biophys Res Commun 298: 714-719. Link: https://goo.gl/M3hjwN
- Fang H, Zhang A, Yu J (2016) Insight into the metabolic mechanism of scoparone on biomarkers for inhibiting Yanghuang syndrome. Scientific Reports 6: 37519. Link: https://goo.gl/M8VZgY
- Keitel V, Donner M, Winandy S, Kubitz R, Häussinger D (2008) Expression and function of the bile acid receptor tgr5 in kupffer cells. Biochemical & Biophysical Research Communications 372: 78-84. Link: https://goo.gl/iqPxi6
- Zhou XH, Zhang AH, Wang L (2016) Novel chinmedomics strategy for discovering effective constituents from ShenQiWan acting on ShenYangXu syndrome.. Chinese Journal of Natural Medicines 14: 561-581. Link: https://goo.gl/6TQwob
- Duboc H, Taché Y, Hofmann AF (2014) The bile acid tgr5 membrane receptor: from basic research to clinical application ☆. Digestive & Liver Disease 46: 302-312. Link: https://goo.gl/4xnb7i
- Nan Y, Zhou X, Liu Q (2016) Serum metabolomics strategy for understanding pharmacological effects of ShenQi pill acting on kidney yang deficiency syndrome. Journal of Chromatography B Analytical Technologies in the Biomedical Life Sciences 1026: 217-226. Link: https://goo.gl/465eA5
- Keitel V, Reich M, Häussinger D (2015) Tgr5: pathogenetic role and/or therapeutic target in fibrosing cholangitis?. Clinical Reviews in Allergy & Immunology 48: 218-225. Link: https://goo.gl/ZHHkjw
- Keitel V, Häussinger D (2013) Tgr5 in cholangiocytes. Current Opinion in Gastroenterology 29: 299-304. Link: https://goo.gl/WzDEzz
- Hodge RJ, Nunez DJ (2016) Therapeutic potential of takeda-g-protein-receptor-5 (tgr5) agonists. hope or hype?. Diabetes Obesity & Metabolism 18: 439-443. Link: https://goo.gl/6PKKvK
- Zhang A, Sun H, Wang X, Jiao G, Yuan Y, et al. (2012) Simultaneous in vivo RP‐HPLC‐DAD quantification of multiple‐component and drug–drug interaction by pharmacokinetics, using 26: 844-850. Link: https://goo.gl/YhGjmx
- Beuers U, Kullakublick GA, Pusl T, Rauws ER, Rust C (2009) Medical treatment of primary sclerosing cholangitis: a role for novel bile acids and other (post-)transcriptional modulators?. Clinical Reviews in Allergy & Immunology 36: 52-61. Link: https://goo.gl/88V2E5
- Wang H, Sun H, Zhang A, Li Y, Wang L, et al. (2013) Rapid identification and comparative analysis of the chemical constituents and metabolites of P hellodendri amurensis cortex and Z hibai dihuang pill by ultra‐performance liquid chromatography with quadrupole TOF‐MS. Journal of separation science 36: 3874-3882. Link: https://goo.gl/y4WgK5
- Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, et al. (2009) Tgr5-mediated bile acid sensing controls glucose homeostasis. Cell Metabolism 10: 167-177. Link: https://goo.gl/ibqaNx
- Zhang A, Sun H, Yan G, Wang P, Wang X (2016) Mass spectrometry‐based metabolomics: applications to biomarker and metabolic pathway research. Biomedical Chromatography 30: 7-12. Link: https://goo.gl/LpMhU3
- Pathak P, Cen X, Nichols RG, Ferrell JM, Boehme S, et al. (2018) Intestine farnesoid x receptor agonist and the gut microbiota activate g‐protein bile acid receptor‐1 signaling to improve metabolism. Hepatology 68: 1574-1588. Link: https://goo.gl/ALM4ws
- Sun H, Zhang A, Yan G, Han Y, Sun W, et al. (2013) Proteomics study on the hepatoprotective effects of traditional Chinese medicine formulae Yin-Chen-Hao-Tang by a combination of two-dimensional polyacrylamide gel electrophoresis and matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Journal of Pharmaceutical and Biomedical Analysis 75: 173-179. Link: https://goo.gl/udPohx
- Wang X, Zhang A, Sun H, Wang P (2012) Systems biology technologies enable personalized traditional Chinese medicine: a systematic review. The American journal of Chinese medicine 40: 1109-1122. Link: https://goo.gl/DRkzos
- Mutlu EA, Gillevet PM, Huzefa R, Masoumeh S, Ammar N, et al. (2012) Colonic microbiome is altered in alcoholism. American Journal of Physiology Gastrointestinal & Liver Physiology 302: 966-978. Link: https://goo.gl/H5NrKG
- Cao H, Zhang A, Zhang H, Sun H, Wang X (2015) The application of metabolomics in traditional Chinese medicine opens up a dialogue between Chinese and Western medicine. Phytotherapy research 29: 159-166. Link: https://goo.gl/Qa31YJ
- Degirolamo C, Rainaldi S, Bovenga F, Murzilli S, Moschetta A (2014) Microbiota modification with probiotics induces hepatic bile acid synthesis via downregulation of the fxr-fgf15 axis in mice. Cell Reports 7: 12-18. Link: https://goo.gl/cD7muQ
- Juste C (2005) Dietary fatty acids, intestinal microbiota and cancer. Bulletin Du Cancer 92: 708-721. Link: https://goo.gl/3irxnK
- Li J, Lu Y, Liu R, Xiong X, Zhang Z, et al. (2011) Dax1 suppresses fxr transactivity as a novel co-repressor. Biochem Biophys Res Commun 412: 660-666. Link: https://goo.gl/TNR8PK
- Zhang A, Sun H, Xu H, Qiu S, Wang X (2013) Cell metabolomics. Omics: a journal of integrative biology 17: 495-501. Link: https://goo.gl/5YKRML
- Chen P, Stärkel P, Turner JR, Ho SB, Schnabl B (2015) Dysbiosis‐induced intestinal inflammation activates tumor necrosis factor receptor i and mediates alcoholic liver disease in mice. Hepatology 61: 883-894. Link: https://goo.gl/NMFhy7
- Inagaki T, Moschetta A, Lee YK, Peng L, Zhao G, et al. (2006) Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci 103: 3920-3925. Link: https://goo.gl/QYS5ZJ
- Devkota S, Wang Y, Musch MW, Leone V, Fehlnerpeach H, et al. (2012) Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in il10-/- mice. Nature 142: 104-108. Link: https://goo.gl/qMwGM2
- Compare D, Coccoli P, Rocco A, Nardone OM, De MS, et al. (2012) Gut--liver axis: the impact of gut microbiota on non alcoholic fatty liver disease. Nutrition Metabolism & Cardiovascular Diseases Nmcd 22: 471-476. Link: https://goo.gl/xvt1sd
- Islam KB, Fukiya S, Hagio M, Fujii N, Ishizuka S, et al. (2012) Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 141: 1773-1781. Link: https://goo.gl/Rqyp6C
- Yu M, Jia HM, Zhou C, Yang Y, Sun LL, et al. (2017) Urinary and fecal metabonomics study of the protective effect of chaihu-shu-gan-san on antibiotic-induced gut microbiota dysbiosis in rats. Sci Rep 7: 46551. Link: https://goo.gl/FX74Px
- Sun H, Wang M, Zhang A, Ni B, Dong H, et al. (2013) UPLC–Q‐TOF–HDMS Analysis of Constituents in the Root of Two Kinds of Aconitum Using a Metabolomics Approach. Phytochemical Analysis 24; 263-276. Link: https://goo.gl/RWQWCE
- Wang F, Meng J, Zhang L, Johnson T, Chen, et al. (2018) Morphine induces changes in the gut microbiome and metabolome in a morphine dependence model. Scientific Reports 3596. Link: https://goo.gl/oVZaU6
- Bernstein H, Bernstein C, Payne CM, Dvorak K (2009) Bile acids as endogenous etiologic agents in gastrointestinal cancer. World Journal of Gastroenterology 15: 3329-3340. Link: https://goo.gl/D31QT7
- Mikó E, Vida A, Kovács T, Ujlaki G, Bai P (2018) Lithocholic acid, a bacterial metabolite reduces breast cancer cell proliferation and aggressiveness. Biochimica et Biophysica Acta (BBA) - Bioenergetics 1859: 958-974. Link: https://goo.gl/1BCpGL
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
