Open Journal of Proteomics and Genomics
miRNA Regulation of Telomerase: A Novel Therapeutic Approach for Cancer
Twinkle Wahi, Sahil Dargan, Sumedha Jaitly and Vibha Rani*
Cite this as
Wahi T, Dargan S, Jaitly S, Rani V (2016) miRNA Regulation of Telomerase: A Novel Therapeutic Approach for Cancer. Open J Proteom Genom 1(1): 013-018. DOI: 10.17352/ojpg.000003Telomeres are repetitive sequence of nucleotides present at the end of the chromosomes. The main function of telomeres is to protect the ends of chromosomes from degradation and fusion. In normal cells, the telomere length decreases after each mitosis cycle, reaching a threshold limit after which the cell undergoes apoptosis. Telomerase is over expressed in approximately 85-90% of the tumors and thus it has become a lucrative target as an anti-cancer therapeutic. In recent past, many therapies targeting telomerase have been pursued, including immunotherapeutic approaches, telomerase targeted gene therapy, and most recently microRNAs telomerase vaccines, making them an attractive target for cancer therapeutics. MicoRNAs (miRNAs) are 20-24 base pair non-coding RNAs that regulate gene expression at post-transcriptional level. miRNAs are important regulators of cancer and can be used to target telomerase, thus acting as a therapeutic for cancer.In this review, we summarize the possibility of broad-spectrum immunotherapy or even immunoprevention and discuss therapeutic approaches based on small molecules to inhibit telomerase activity. Also focusing on microRNA and how it controls the expression of telomerase to prevent tumor development.
Abbreviations
CTL: Cytotoxic T-cell; APC: Antigen Presenting Cells; DC: Dendritic Cells; PTEN: Phosphatase and Tensin homolog; AGO2: Argonaute 2: RISC: Catalytic Component: TRF-1: Telomere Repeat Factor; POT1: Protection of Telomerase 1; RAP1: Repressor/Activator Orotein; TRP2: Telomere Repeat Factor 2; TIN2: TRF1 and TRF2: Interacting Nuclear Protein 2; PI3K: Phosphoinositide 3-kinase; HNSCC: Head and Neck Squamous Cell Carcinoma cells; PITX1: Ducer and Activator of Transcription 3 (STAT3); ADAM9 (ADAM Metallopeptidase Domain 9); Non-small cell lung cancer (NSCLC)
Introduction
Telomere and telomerase biology
Telomere refers to the repetitive sequence of nucleotides present at the end of the chromosomes, having a structure which is different from the rest of the chromatin [1]. Telomeres consist of short and repeating sequence of d [TTAGGG] with a single stranded 3’ overhang that forms a loop like structure and invades back into the duplex of telomere [2]. The main function of telomeres is capping the end of the chromosomes to prevent them from degradation and fusion. Protective function of telomere is associated with a group of telomere associated proteins known as sheltrin complex. The complex comprises of protection of telomerase 1(POT1), repressor/activator protein 1(RAP1), telomere repeat factor 1(TRF1), telomere repeat factor 2(TRP2), TRF1 and TRF2 –interacting nuclear protein 2(TIN2) and TINT1/PTOP/PIP1 protein (TPP1) [3,4]. POT1 binds to the single stranded overhangs whereas TRF1 and TRF2 bind to the double stranded DNA [5-7]. TIN2 binds TRF1 and TRF2 through protein interactions with distinct domains.TPP1 interacts with TIN1 and POT1 and forms a complex [8-10]. RAP1, TRF1- interacting protein 2 (TIN2), and TPP1 associate with these DNA-binding proteins to form a functional complex telosome. TIN2 is a key component of the telosome, and associates with both TRF1 and TRF2 and is essential for bringing together the DNA-binding proteins within the telosome complex. TRF1, TRF2 and POT1 have a key role in telomere protection by suppressing the activation of the DNA damage signaling and repair pathways at chromosome ends [11-13].
Shortening of the telomere takes place due to a phenomenon known as end replication problem. Phenomenon characterized by the shortening of the 3’ end of DNA with each cell division as DNA polymerase cannot completely replicate the strand [14,15]. At a particular threshold of telomere attenuation the damage repair system activates the p53 or the p16INK4a signaling pathway which initiates apoptosis. A various other environmental and genetic factors can cause telomere damage of increased shortening of telomere length. These include stress, inflammation, ROS generation and smoking [16].
Telomerase is an enzyme which maintains the length of the telomeres. The human telomerase is a holoenzyme and consist of 2 subunits: hTERT (Human telomerase reverse transcriptase), responsible for synthesis of DNA from RNA template and hTR (RNA template), which acts as a template for the addition of nucleotide sequences [17]. hTERT utilizes the template region (3’-CAAUCCCAAUC-5’) of TERC to add TTAGGG DNA repeats and thereby extends single stranded 3’ telomeric strands [18]. Elongation of telomere by telomerase leads to stabilization of the chromosome and thus high telomerase activities observed in progenitor and stem cells [19]. However irregular induction of telomerase may cause cancer development. Moreover, 90% of the cancer cells express telomerase to elongate the lengths of the telomere. The activity of telomerase is highly up regulated in many tumors including, colon [20,21], breast [22,23], ovarian [24], oral [25,26], pancreas [27], melanoma[28] and soft tissue cancers [29].
MicroRNAs: Novel class of regulator for gene silencing
MicoRNA’s are 20-24 base pair non-coding RNA’s that regulate gene expression at post-transcriptional level. They influence a number of tumor related processes such as apoptosis, differentiation, proliferation and migration. The transcription of miRNAs is carried out by RNA Polymerase II forming a hairpin like primary transcript known as pri-miRNA. In nucleus, pri-miRNA is refined to a precursor miRNA called pre-miRNA by an RNase III enzyme called Drosha. Pre-miRNA is then subsequently exported to the cytoplasm by Exportin-5 protein which is one of the Ran dependent nuclear transport receptor families [30]. TRBP and DICER cleavepre-miRNA to produce single stranded mature mi-RNA and incorporate into RNA induced silencing complex (RISC), containing GW182 and AGO2 proteins. As a part of RISC, mature mi-RNA interacts with complementary sequences in 3’ UTR’s causing mRNA degradation or translation inhibition thereby playing a role in gene regulation (Figure 1) [31].
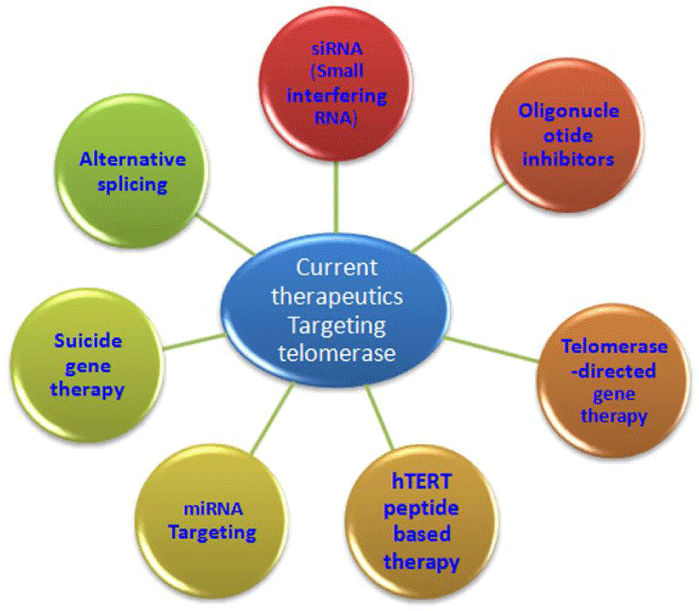
Current therapeutics targeting telomerase
Due to the high complexity of human telomerase various therapeutics targeting telomerase have been developed. The inhibition of hTERT is a good option as it is up regulated in most of the cancer malignancies and is the rate-limiting subunit of the telomerase complex, is therefore an attractive target for cancer vaccination. Furthermore, since somatic cells exhibit no or low levels of telomerase, it can be targeted. The following are the therapeutics which target telomerase (Figure 2).
hTERT peptide based therapy: Active immunotherapy involves the use of tumor vaccines to elicit endogenous antitumor immune responses, hence “educating” immune cells to produce a long-lasting effect [32]. Immunotherapy targeting hTERT is mainly focused to elicit hTERT-specific T lymphocyte immune responses, especially CTL responses that can specifically kill hTERT+ tumor cells. Here DCs (dendritic cells) which act as APC (antigen presenting cells) are prepared ex vivo and transfected with hTERT/tumor RNA or transduced with viral vector expressing hTERT peptides [33]. After immunization, DCs migrate to draining lymph node where DCs induce hTERT-specific CD4+ TH cells and CD8+ CTLs to trigger T-cell immunity against tumor cells. Direct injection of hTERT peptides or viral vectors expressing hTERT peptides induce inflammation and recruit immature DCs to capture expressed TAAs. The DCs mature and migrate to draining lymph node where they induce hTERT-specific CD4+ and CD8+ T cells to trigger T-cell immunity against hTERT+ tumor cells.
Telomerase-directed gene therapy: The telomerase gene promoters can be used to target therapeutic genes in cancer cells as a consequence of tumor-specific gene expression of telomerase. Telomerase gene therapy is a versatile, powerful, and useful strategy which has the advantage of causing immediate cancer cell death. The promoters of telomerase in cancer cells are targets for a tumor specific gene therapy that selectively kills cancer cells and leaves normal cells unharmed by expressing high concentrations of a therapeutic protein [34].
Oligonucleotide inhibitors: Antisense oligonucleotides and chemically-modified nucleic acids have been shown to be associated with onset of senescence and/or apoptosis [35-38]. Targets include the RNA template, hTERT protein, and associated proteins. As an example imetelstat (GRN163L) targets the RNA template for hTERT by binding to the catalytic site of telomerase [39]. Imetelstat is a 13-mer (5’-TAGGGTTAGACAA-3’) that is complementary to nine nucleotides in the template region and four nucleotides 5’ of the template region of TERC [40].
siRNA(Small interfering RNA): In this response, RNAs of 21-23 nucleotides in length called siRNA, are snipped from longer dsRNA, by an enzyme called dicer. The antisense strand is used by a RNA inducing silencing complex (RISC) and guide mRNA cleavage of target gene. siRNA mediated silencing of telomerase have been witnessed in literature as a therapy against many cancers. SiRNA mediated inhibition of telomerase is reported to enhance the anticancerorous potential of drugs (doxorubicin and others) in various cancerous models [41].
Alternative splicing: The hTERT gene of 42 kb on human chromosome at 5p15.33 contains 16 exons and can be spliced into multiple isoforms [42]. Alternatively spliced 22 isoforms including minus alpha, minus beta, or both (minus alpha beta) do not contain reverse transcriptase activity and hence they cannot elongate telomeres [43, 44].The minus alpha splicing isoform uses an alternative 3’ splice acceptor site 36 bp into exon 6, resulting in an in frame transcript that is translated into a dominant-negative protein without reverse transcriptase activity [45,46]. Whereas minus beta splicing isoform skips exons 7 and 8, creating a frame-shift that further leads to a premature stop codon in exon 10.
Suicide gene therapy: In this strategy, an adenoviral system constructed by engineering the bacterial nitroreductase (NTR) through highly active TERT/TERC promoter in tumor cells is exploited to produce cytotoxic end products [47]. NTR produced by TERT/TERC active cancer cells further converts the pro-drug CB1954 into active cytotoxic 2- and4-hydroxylamino derivatives that form DNA crosslinks via an N-acetoxy intermediate. This approach induced significant tumor reduction in xenograft models and cell death in various cancer cell lines [48].
Regulation of telomerase by miRNA
miRNAs bind to the 3’ UTR of the target mRNA and interfere with the protein production by destabilizing the target mRNA. For example, miR-138 is reported to directly bind to the target site in hTERT 3’ UTR and repress the expression of protein [49]. A study also demonstrates that miRNA’s- let-7g*, miR-133a, miR-342-5p, miR-491-5p, and miR-541-3p also regulate hTERT directly [50]. Though, the exact mechanism of all of the miRNA’s is still unknown. The following miRNAs have known to regulate telomerase, and can be examined for future microRNA based therapeutics:
miR-135b: Reported to promote tumor transformation and progression in colon cancer prostate cancer, pancreatic ductal adenocarcinoma, anaplastic large-cell lymphomas [51]. The expression level of miR-135b is tightly regulated by tissue- specific micro-environment niches during the MSC fate-determination. These observations support the oncogenic role of miR-135b on cancer progression such that it may serve as a biomarker for cancer.
miR-21: PTEN(phosphatase and tensin homolog deleted on chromosome 10)function as a tumor suppressor and its decreased expression results in activation of PI3K/AKT which enhances tumor progression. Various studies have showed that mir-21 down regulates PTEN. Mir-21 expression inhibited apoptosis and induced proliferation accompanied by the negative regulation of s hTERT expression via the phosphoinositide 3-kinase (PI3K) signaling pathway by binding to 3’ UTR of PTEN leading to inhibition of its translation .moreover the genes pAKT and PI3K, downstream of Htert were upregulated by mir-21in Hypertrophic Scar Fibroblasts [52].
miR-19b: PITX1(Paired Like Homeodomain 1) mRNA is a direct target of miR-19b and the down regulation of PITX1 by miR-19b ultimately induces enhancement of hTERT mRNA expression [53]. PITX1 (paired like homeodomain 1) is a tumor suppressor gene as it binds directly to the promoter and directly inhibits hTERT gene transcription thereby resulting in the inhibition of telomerase activity and cell -proliferation [54]. It promotes PI3K pathway signaling through inhibition of PTEN (Phosphatase and tensin homolog) expression [55, 56].miR-19b is included in the miR-17-92 and the miR-106-363 clusters. These clusters carry out pleiotropic functions during both normal development and malignant transformation, as they act to promote proliferation and sustain cell survival [57,58].
miR- 346: this mediates the upregulation of hTERT gene expression in human cervical cancers. The middle sequence motif (nt 8–13, CCGCAU) of miR-346is responsible for binding to G rich sequence binding factor 1(GRSF-1) and forms a bulge loop .This binding recruits hTERT mRNA to ribosomes for translation in an AGO2-independent manner.mir-346 enhanced proliferation of cervical tissue cells by specifically targeting the 3’ UTR f hTERT transcript [59].
smiR-138: This mediates the suppression of hTERT expression in cervical cancers in an AGO2(Argonaute 2) dependent manner by binding to 3’ UTR of cancer cell lines. miR 138 and miR 346 binding sites (nt 20 to 34) are in a common region of the hTERT 3’UTR for GRSF1(G- rich RNA sequence binding factor 1) overlap by 9 bases and hence they competes for binding to sites within the hTERT 3’ UTRmRNA expression correlate with the ratio of miR-346 to miR-138. GRSF1 serve as universal mediator to regulate gene expression which facilitates the recruitment of the target gene mRNA to ribosomes for translation [59].
miR-512-5p: The study by s Jun Li et al. showed that over expression of this miRNA in HNSCC(Head and Neck Squamous Cell Carcinoma cells),suppressed tumor growth in vivo and cell proliferation through elevated apoptosis, inhibition of telomerase activity, decrease of telomere binding proteins and shortening of telomere length by targeting hTERT down regulation in vitro. Mir512-5p serve as a tumor suppressor and a direct target of hTERT. This might serve as a therapeutic agent in miRNA based HNSCC therapy [60].
miR-155: act as key regulator in breast cancer cell expression. It is efficiently upregulated with reduced TRF1 protein levels. Mediate telomere elongation, increased telomere fragility and chromosome instability by reducing the expression of TRF1 (telomere repeat factor 1) i.e. a part of shelterin complex. In contrast reducing the expression of mir-155 ensures genomic stability with improved telomere function it directly controls TRF1 expression by binding to its 3’ UTR leading to translational repression [61-64].
miR- 29: Acts as a tumor suppressor in lung tumors where it targets both DNMT (DNA methyltransferase) 3A and 3B and restores its normal dna methylation patterns, as well as it also induces re expression of tumor suppressor genes such as FHIT and WWOX and inhibit tumorigenicity [65]. In breast cancer, overexpression of mir-29a suppressed the expression of TTP (tristetraprolin), a protein responsible for the degradation of m-RNA in cooperation with oncogenic Rassignaling [66].
miRNAs in cancer diagnosis and therapy
Various therapeutics strategies have been developed involving re-introduction of miRNAs lost in cancer or inhibition of oncogenic miRNAs. For the inhibition of pro-oncogenic miRNA therapeutic treatments involve intravenous injections with cholesterol-conjugated 2’O-methyl which are inhibitors ofmiR-10b, over expressed in metastatic breast cancer [67-68]. Similarly the therapeutic potential of reintroducing miR-26a in mouse model for hepatocellular carcinoma, where systemic delivery of miR-26a expressed from an adeno-associated viral vector resulted in inhibited cancer cell proliferation [69].
Current perspectives
miRNA is believed to regulate the expression of multiple targets and many miRNAs have been linked to tumorgenesis, differentiation, immune system and cell death. It was found that high level of telomerase activity is expressed in immortal cells and in 85-90% of human cancers. Dysregulation of microRNAs (miRNAs) plays a critical role in tumor growth and progression and thus a focus of attention against cancer treatment. Transfection with miR-539 mimic significantly coupled with reduced expression of anti-apoptotic proteins Bcl-2 and Bcl-xL and decreased phosphorylation of Stat3 [70]. Other than this delivery of miR-590 mimic was found to decrease endogenous ADAM9 expression in Non-small-cell lung carcinoma (NSCLC) cells. Enforced expression of a miRNA-resistant form of ADAM9 significantly restored the aggressive behaviors in miR-590-overexpressing NSCLC cells. Restoration of miR-590 may provide a promising therapeutic strategy for NSCLC [71]. Telomerase regulation can also be target of cancer treatment as the restoration of miR-29a may be a rational therapeutic strategy for the treatment of hTERT-mediated gastric cancer metastasis [72]. The repression of telomerase and shorter telomeres in humans act as an anticancer protection mechanism. Although there is still much to understand about the regulation of telomerase, it remains a very attractive and novel target for cancer therapeutics.
Conclusion
In this review, we have summarized the current therapeutics targeting telomerase, especially focusing on microRNA. Telomerase serves as a lucrative target as it is up-regulated in approximately 90% of the tumors. In addition, the current treatments of cancer available in the field of medicine have a lot of side effects thus promoting the development of alternative ways to ameliorate cancer. Future research needs to be done to unveil the pathways involved in the regulation of telomerase-miRNA expression and to link telomerase-miRNA to key biological processes related to cancer. Unlike other small-RNAs, miRNAs do not require perfect base pairing, and thus, can regulate a network of broad, yet specific, genes and can be targeted as therapeutic gent against various disease. Future research must be into the design and application of efficient synthetic systems for miRNA delivery. Overall, a better understanding of the rational design of miRNA delivery systems will promote their translation into effective clinical treatments. Evidence suggests that miRNA-based therapies, either restoring or repressing expression and activity, hold great promise. However, Critical hurdles often involving delivery of miRNA-targeting agents remain to be overcome before transition to clinical applications.
- Giraud-Panis MJ, Pisano S, Poulet A, Le Du MH, Gilson E (2010) Structural identity of telomeric complexes. FEBS Lett 584: 3785-3799. Link: https://goo.gl/RwoYFf
- To-Miles FYL, Backman CL (2016) What telomeres say about activity and health: A rapid review. Can J Occup Ther pii: 0008417415627345. Link: https://goo.gl/tQ3ep5
- Fyhrquist F, Saijonmaa O, Strandberg T (2013) The roles of senescence and telomere shortening in cardiovascular disease. Nat RevCardiol 10: 274–283. Link: https://goo.gl/CnCNBG
- Griffith JD,Comeau L, Rosenfield S, Stansel RM, Bianchi A, et al. (1999) Mammalian telomeres end in a large duplex loop. Cell 97: 503–514. Link: https://goo.gl/8PHNBO
- De Lange, Shelterin T (2005) The protein complex that shapes and safeguards human telomeres. Genes Dev 19: 2100–2110. Link: https://goo.gl/bdKyzo
- Loayza D, de Lange T (2003) POT1 as a terminal transducer of TRF1 telomere length control. Nature 423: 1013–1018. Link: https://goo.gl/Bz3plQ
- Broccoli D, Smogorzewska A, Chong L, de-Lange T (1997) Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat Genet 17: 231–235. Link: https://goo.gl/k3KeMB
- Bianchi A, Stansel RM, Fairall L, Griffith JD, Rhodes D, et al. (1999) TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J 18: 5735–5744. Link: https://goo.gl/dLTGEs
- Ye JZ, Donigian JR, van Overbeek M, Loayza D, Luo Y, et al. (2004) Tin2 binds trf1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J Biol Chem 279: 47264–47271. Link: https://goo.gl/1LdR3R
- Frescas D, de Lange T (2014) Binding of TPP1 protein to TIN2 protein is required for POT1a, b protein-mediated telomere protection. J Biol Chem 289: 24180–24187. Link: https://goo.gl/n3nsZl
- Liu D, Safari A, O’Connor MS, Chan DW, Laegeler A, et al. (2004) Ptop interacts with POT1 and regulates its localization to telomeres. Nat Cell Boil 6: 673–680. Link: https://goo.gl/hN2HOx
- Denchi EL, de Lange T (2007) Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448: 1068-1071. Link: https://goo.gl/uEIIb2
- Martínez P, Thanasoula M, Muñoz P, Liao C, Tejera A, et al. (2009) Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev 23: 2060-2075. Link: https://goo.gl/hLDkE5
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, et al. (2009) Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell138: 90-103. Link: https://goo.gl/sgOss3
- Boccardi V, Herbig U (2012) Telomerase gene therapy: A novel approach to combat aging. EMBO Mol Med 4: 685–687 Link: https://goo.gl/YPcxCX
- Artandi SE, DePinho RA (2010) Telomeres and telomerase in cancer. Carcinogenesis 31: 9–18. Link: https://goo.gl/fkT4If
- Nawrot TS, Staessen JA, Holvoet P, Struijker-Boudier HA, Schiffers P, et al. (2010) Telomere length and its associations with oxidized-LDL, carotid artery distensibility and smoking. Front Biosci 2: 1164–1168. Link: https://goo.gl/8K89iR
- Zhu H, Blecher M, van der Harst P (2011) Healthy aging and disease: Role for telomere biology? ClinSci 120: 427–440. Link: https://goo.gl/APQf8g
- Morin GB (1989) The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell 1989; 59: 521–529. Link: https://goo.gl/N44wDv
- Okayasu I, Mitomi H, Yamashita K, Mikami T, Fujiwara M, et al. (1998) Telomerase activity significantly correlates with cell differentiation, proliferation and lymph node metastasis in colorectal carcinomas. J Cancer Res Clin Oncol 124: 444–449. Link: https://goo.gl/mUSx8K
- Yoshida R, Kiyozuka Y, Ichiyoshi H, Senzaki H, Takada H, et al. (1989) Change in telomerase activity during human colorectal carcinogenesis. Anticancer Res 19: 2167–2172. Link: https://goo.gl/j4TjWl
- Carey LA, Kim NW, Goodman S, Marks J, Henderson G, et al. (1999) Telomerase activity and prognosis in primary breast cancers. J ClinOncol 17: 3075–3081. Link: https://goo.gl/OlfVaM
- Poremba C, Heine B, Diallo R, Heinecke A, Wai, D, et al. (2002) Telomerase as a prognostic marker in breast cancer: High-throughput tissue microarray analysis of hTERT and HTR. J Pathol 198: 181–189. Link: https://goo.gl/q5D6GM
- Oishi T, Kigawa J, Minagawa Y, Shimada M, Takahashi M, et al. (1998) Alteration of telomerase activity associated with development and extension of epithelial ovarian cancer. Obstet Gynecol91: 568–571. Link: https://goo.gl/uhXwg0
- Miyoshi Y, Tsukinoki K, Imaizumi T, Yamada Y, Ishizaki T, et al. (1999) Telomerase activity in oral cancer. Oral Oncol 35: 283–289. Link: https://goo.gl/I2CGjh
- Mori K, Sato S, Kodama M, Habu M, Takahashi O, et al. (2013) Oral cancer diagnosis via a ferrocenylnaphthalenediimide-based electrochemical telomerase assay. Clin Chem59: 289–295. Link: https://goo.gl/L7hQEk
- Hata T, Ishida M, Motoi F, Yamaguchi T, Naitoh T, et al. (2016) Telomerase activity in pancreatic juice differentiates pancreatic cancer from chronic pancreatitis: A meta-analysis. Pancreatology 16: 372-381. Link: https://goo.gl/byHGiJ
- Pirker C, Holzmann K, Spiegl-Kreinecker S, Elbling L, Thallinger C, et al. (2003) Chromosomal imbalances in primary and metastatic melanomas: Over-representation of essential telomerase genes. Melanoma Res 13: 483–492. Link: https://goo.gl/mDXSHd
- Tomoda R, Seto M, Tsumuki H, Iida K, Yamazaki T, et al. (2002) Telomerase activity and human telomerase reverse transcriptase mRNA expression are correlated with clinical aggressiveness in soft tissue tumors. Cancer 95: 1127–1133. Link: https://goo.gl/z2qBth
- Sarin KY, Cheung P, Gilison D, Lee E, Tennen RI, et al. (2005) Conditional telomerase induction causes proliferation of hair follicle stem cells. Nature 436: 1048–1052. Link: https://goo.gl/tncjkF
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U (2004) Nuclear export of microRNA precursors. Science 303: 95–98. Link: https://goo.gl/97rAFY
- Iorio MV, Croce CM (2012) MicroRNA dysregulation in cancer: Diagnostics, monitoring andtherapeutics. A comprehensive review. EMBO Mol Med 4: 143–159. Link: https://goo.gl/c1AKvl
- Shaw DR, LoBuglio AF (2005) Genetic immunotherapy approaches. In: Cancer Gene TherapyCuriel DT, Douglas JT, eds. Totowa, New Jersey: Humana Press Inc 129-141. Link: https://goo.gl/F4b4Fd
- Longfei H, Janice WST, Junjian H, Peitang H, Cuifen H, et al. (2006) Cancer Immunotherapy Targeting the Telomerase Reverse Transcriptase. Cell Mol Immunol 3: 1-9. Link: https://goo.gl/V74w7t
- Nemunaitis J, Tong AW, Nemunaitis M, Senzer N, Phadke AP, et al. (2010) A phase I study of telomerase-specific replication competent oncolytic adenovirus (telomelysin) for various solid tumors. Mol Ther 18: 429-434. Link: https://goo.gl/ma7rUi
- Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, et al. (1995) The RNA component of human telomerase. Science 269: 1236-1241. Link: https://goo.gl/8XkYCj
- Norton JC, Piatyszek MA, Wright WE, Shay JW, Corey DR (1996) Inhibition of human telomerase activity by peptide nucleic acids. Nat Biotechnol1996; 14: 615–619. Link: https://goo.gl/7yllOC
- Herbert B, Pitts AE, Baker SI, Hamilton SE, Wright WE, et al. (1999) Inhibition of human telomerase in immortal human cells leads to progressive telomere shortening and cell death. Proc Natl Acad Sc USA 96: 14276–14281. Link: https://goo.gl/ZeQjqI
- Kondo S, Tanaka Y, Kondo Y, Hitomi M, Barnett GH, et al. (1998) Antisense telomerase treatment: Induction of two distinct pathways, apoptosis and differentiation. FASEB J 12: 801–811. Link: https://goo.gl/SjRy7G
- Harley CB (2008) Telomerase and cancer therapeutics. Nat Rev Cancer 8: 167–179. Link: https://goo.gl/42QJzL
- Xuejun D, Anding L, Cindy Z, Jianguo F, Zhuan Z, et al. (2009) SiRNA inhibition of telomerase enhances the anti-cancer effect of doxorubicin in breast cancer cells. BMC Cancer 9: 133. Link: https://goo.gl/akXhX3
- Andrzej K, David DLB, Helen EA, Gary RH, Deon JV, et al. (1997) Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum Mol Genet 6: 2011–2019. Link: https://goo.gl/zdRpVK
- Radmila H, Jiří N, Henry RB (2012) Alternatively spliced telomerase reverse transcriptase variants lacking telomerase activity stimulate cell proliferation. Mol CellBiol 32: 4283–4296. Link: https://goo.gl/3qoPDW
- Saeboe-Larssen S, Fossberg E, Gaudernack G (2006) Characterization of novel alternative splicing sites in human telomerase reverse transcriptase (hTERT): analysis of expression and mutual correlation in mRNA isoforms from normal and tumour tissues. BMC MolBiol 7: 26 41. Link: https://goo.gl/fc6x9u
- Yi X, White DM, Aisner DL, Baur JA, Wright WE, et al. (2002) An alternate splicing variant of the human telomerase catalytic subunit inhibits telomerase activity. Neoplasia 2: 433–440. Link: https://goo.gl/eW3x7n
- Colgin LM, Wilkinson C, Englezou A, Kilian A, Robinson MO, et al. (2000) The Htert alpha splice variant is a dominant negative inhibitor of telomerase activity. Neoplasia 2: 426–432. Link: https://goo.gl/LAK1yK
- Wong MS, Wright WE, Shay JW (2014) Alternative splicing regulation of telomerase: a new paradigm? Trends in Genetics 30: 430-438. Link: https://goo.gl/o9g4cJ
- Bilsland AE, Anderson CJ, Fletcher-Monaghan AJ, McGregor F, Evans TR, et al. (2003) Selective ablation of human cancer cells by telomerase-specific adenoviral suicide gene therapy vectors expressing bacterial nitroreductase. Oncogene 22: 370–380. Link: https://goo.gl/Gf4mmQ
- Mitomo S, Maesawa C, Ogasawara S, Iwaya T, Shibazaki M, et al. (2008) Down regulation of miR-138 is associated with overexpression of human telomerase reverse transcriptase protein in human anaplastic thyroid carcinoma cell lines. Cancer Sci 99: 280–286. Link: https://goo.gl/LU2wJO
- Hrdlicˇkova´ R, Nehyba J, Bargmann W, Bose HR Jr. (2014) Multiple Tumor Suppressor microRNAs Regulate Telomerase and TCF7, an Important Transcriptional Regulator of the Wnt Pathway. PLoS ONE 9: e86990. Link: https://goo.gl/AeoUkV
- Plumb JA, Bilsland A, Kakani R, Zhao J, Glasspool RM, et al. (2001) Telomerase-specific suicide gene therapy vectors expressing bacterial nitroreductase sensitize human cancer cells to the pro-drug CB1954. Oncogene 20: 7797–7803. Link: https://goo.gl/dZ11nq
- Hua-Yu Z, Chao L, Wen-Dong B, Lin-Lin S, Jia-Qi L, et al. (2014) MicroRNA-21 regulates hTERT via PTEN in hypertrophic scar fibroblasts. PLoS One 9: 97114. Link: https://goo.gl/jKt1tI
- Vanhaesebroeck B, Stephens L, Hawkins P (2012) PI3K signalling: the path to discovery and understanding. Nat Rev Mol Cell Biol 13: 195–203. Link: https://goo.gl/EukVYZ
- Takahito O, Sunamura N, Yuji N, Mitsuhiko O, Futoshi, et al. (2015) miR-19b regulates hTERT mRNA expression through targeting PITX1 mRNA in melanoma cells. Scientific Rep 5: 8201. Link: https://goo.gl/CClyE8
- Qi DL, Ohhira T, Fujisaki C, Inoue T, Ohta T, et al. (2011) Identification of PITX1 as a TERT suppressor gene located on human chromosome 5. Mol Cell Biol 31: 1624–1636. Link: https://goo.gl/CIruCH
- Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, et al. (2009) miR-19 is a key oncogenic component ofmir-17-92. Genes Dev 23: 2839–2849. Link: https://goo.gl/XaDBIj
- Mavrakis KJ, Wolfe AL, Oricchio E, Palomero T, de Keersmaecker K, et al. (2010) Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat Cell Biol 12: 372–379. Link: https://goo.gl/gvdBe3
- Olive V, Jiang I, He L (2010) mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol 42: 1348–1354. Link: https://goo.gl/x3MeHP
- Ge Song, Renjie Wang, Junfei Guo, Xuyuan Liu, Fang Wang, et al. (2015) miR-346 and miR-138competitively regulate hTERTin GRSF1- and AGO2-dependentmanners, respectively. Scientific reports 5: 15793. Link: https://goo.gl/ndItk9
- Jun Li, Han Lei, Yong Xu, Ze-zhang Tao (2015) miR-512-5p Suppresses Tumor Growth by Targeting hTERT in Telomerase Positive Head and Neck Squamous Cell Carcinoma In Vitro and In Vivo. PLOS One 10: e0135265. Link: https://goo.gl/pjoflR
- Muñoz P, Blanco R, de Carcer G, Schoeftner S, Benetti R, et al. (2009) TRF1 controls telomere length and mitotic fidelity in epithelial homeostasis. Mol Cell Biol 29: 1608-1625. Link:© https://goo.gl/16gmUE
- Ancelin K, Brunori M, Bauwens S, Koering CE, Brun C, et al. (2002) Targeting assay to study the cis functions of human telomeric proteins: evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2. Mol Cell Biol 22: 3474-3487. Link: https://goo.gl/5U4YTO
- Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, et al. (2000) Control of human telomere length by TRF1 and TRF2. Mol Cell Biol 20: 1659-1668. Link: https://goo.gl/atbFxh
- Roberto Dinami, Cristiana Ercolani, Eleonora Petti, Silvano Piazza, YariCiani, et al. (2014) miR-155 drives telomere fragility in human breast cancer by targeting TRF1.Cancer Res 74: 4145-4156. Link: https://goo.gl/eYr4E8
- Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, et al. (2007) MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA 104: 15805-15810. Link: https://goo.gl/VwFjxO
- Gebeshuber CA, Zatloukal K, Martinez J (2009) miR-29a suppresses tristetraprolin, which is a regulator of epithelial polarity and metastasis. EMBO Rep 10: 400-405. Link: https://goo.gl/2xsRHL
- Ma L, Teruya-Feldstein J, Weinberg RA (2007) Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449: 682-688. Link: https://goo.gl/x2ikQB
- Ma L, Reinhardt F, Pan E, Soutschek J, Bhat B, et al. (2010) Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat. Biotech 28: 341-347. Link: https://goo.gl/47pwSr
- Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, et al. (2009) Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 137: 1005-1017. Link: https://goo.gl/vBlncJ
- Zhu C, Zhou R, Zhou Q, Chang Y, Jiang (2016) MmicroRNA-539 suppresses tumor growth and tumorigenesis and overcomes arsenic trioxide resistance in hepatocellular carcinoma. Life Sci 166: 34-40. Link: https://goo.gl/2oCxrX
- Wang FF, Wang S, Xue WH, Cheng JL (2016) microRNA-590 suppresses the tumorigenesis and invasiveness of non-small cell lung cancer cells by targeting ADAM9. Mol Cell Biochem 423: 29-37. Link: https://goo.gl/m1WnNr
- He B, Yu-Feng X, Bo T, Yu-Yun W, Chang-J Hu, et al. (2016) hTERT mediates gastric cancer metastasis partially through the indirect targeting of ITGB1 by microRNA-29a. Sci Rep 6: 21955. Link: https://goo.gl/Nv96Zo
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
