Open Journal of Cell and Protein Science
Protein folding, misfolding, and coping mechanism of cells–A short discussion
K Geetha Roshni*
Cite this as
Roshni KG (2021) Protein folding, misfolding, and coping mechanism of cells–A short discussion. Open J Cell Protein Sci 4(1): 001-004. DOI: 10.17352/ojcps.000003Inroduction
Proteins are not rigid structures, they are long polypeptide chains of ~50 or more “residues”. Proteins are generally thought to adopt unique structures determined by their amino acid sequences. However, Proteins are not strictly static objects, but rather populate ensembles of (sometimes similar) conformations. Transitions between these states occur on a variety of length scales (tenths of Å to nm) and time scales (ns to s), and have been linked to functionally relevant phenomena such as enzyme catalysis [1] and allosteric signaling [2].
The study of protein dynamics is most directly concerned with the transitions between these states, but can also involve the nature and equilibrium populations of the states themselves. These two perspectives—kinetics and thermodynamics, respectively—can be conceptually synthesized in an “energy landscape” paradigm: [3] highly populated states and the kinetics of transitions between them can be described by the depths of energy wells and the heights of energy barriers, respectively.
Protein Science is a complete study of Proteins, their structure, function and influence as enzymes, etc. Understanding protein function requires detailed knowledge about protein dynamics, i.e. the different conformational states the system can adopt.
Protein dynamics and protein folding
In this mini-review, we will discuss about protein dynamics and protein folding, misfolding and its causes.
Proteins have shapes and stable structures. Their dynamics include binding to other molecules such as Proteins, DNA and metabolites. They catalyze reactions (enzymes) and cause movement such as muscle contractions, rotary motors, etc.
In Ligand binding: Proteins can change their shape. Protein structures fluctuate on many different timescales. They can unfold and refold. They can switch to another conformation (Lock and Key; Induced Fit models).
When Proteins come off the ribosome, they typically fold. Proteins are folded structures and are held together by various forms of molecular interactions. The molecular interactions include the hydrophobic interactions, disulfide bonds formed in the Proteins, and the thermodynamic stability of the complex.
Protein folding is a process by which a polypeptide chain folds to become a biologically active protein in its native 3D structure. Protein structure is important and is linked to its function. Folded Proteins are held together by various molecular interactions. What cause the Proteins To fold is understood by their thermodynamics, but how they fold is explained by protein folding kinetics.
Four forces driving protein structure formation such as Hydrophobic effects, Vander Waals forces, H-bonds, hydrostatic interactions.
Individual classes of interactions can be strongly energetically favorable or strongly energetically unfavorable.
A. To fold, the favorable interactions include
I. Enthalpy from Vander Waals packing interactions
II. Hydrophobic effect (H2O entropy)
III. Gain of protein-protein H bonds
IV. Electrostatic effects
B. To Not Fold, the unfavorable interactions include
I. Protein conformational entropy
II. Loss of protein-water H-bonding [4].
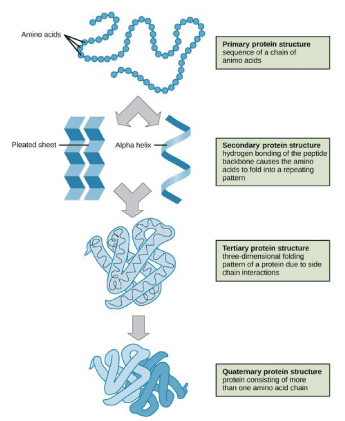
➢ Protein folding occurs in four stages namely:
1. Primary Structure
2. Secondary Structure
3. Tertiary Structure
4. Quaternary Structure.
Protein structure
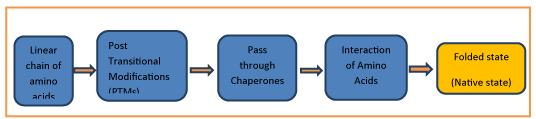
The wide variety of 3-dimensional protein structures corresponds to the diversity of functions Proteins fulfil. With all the above knowledge and criteria of knowing how a protein folds, Figure 1 explains it briefly.
Proteins fold in three dimensions. Protein structure is organized hierarchically from so-called primary structure to quaternary structure. Higher-level structures are motifs and domains.
How a protein folds
Above all the wide variety of conformations is due to the huge amount of different sequences of amino acid residues. The primary structure is the sequence of residues in the polypedptide chain.
Secondary Structure is a local regularly occurring structure in Proteins and is mainly formed through hydrogen bonds between backbone atoms. So-called random coils, loops or turns do not have a stable secondary structure. There are two types of stable secondary structures, the Alpha helices and the beta-sheets. Alpha-helices involves intramolecular hydrogen bonding. In an alpha helix, the carbonyl (C=O) of one amino acid is hydrogen bonded to the amino H (N-H) of an amino acid that is four down the chain. The helical structure which resembles a curled ribbon is caused by the pattern of bonding that pulls the polypeptide chain with each turn of the helix containing 3.6 amino acids. They are free to interact, as the R groups of the amino acids stick out from the alpha helices. These alpha helices are formed when the R group is large. In a Beta-sheet, two or more segments of a polypeptide chain line up next to each other, forming a sheet-like structure held together by hydrogen bonds. Hydrogen bonds form between the carbonyl and amino groups of backbone, meanwhile the R groups extend both above and below the plane of the sheet. Beta-sheets may be parallel strands which point in the same direction i.e. that their N- and C-terminal match up. They can be antiparallel which are pointing in the opposite directions i.e. their N-terminus of one strand id positioned opposite and next to the C-terminus of the other. These beta-sheets are formed when the size of the R group is small to moderate [5]. Alpha-helices and beta-sheets are preferably located at the core of the protein, whereat loops prefer to reside in outer regions.
Tertiary structure expresses the packing of alpha-helices, beta-sheets and random coils with respect to each other on the level of one whole polypeptide chain.
Quaternary structure only exists, if there is more than one polypeptide chain present in a complex protein. Then quaternary structure expresses the spatial organization of the chains [6]. These stages are explained clearly in a schematic manner in Figure 2.
Example of quaternary structure - haemoglobin or DNA polymerase.
Factors influencing protein folding
Protein folding is a very sensitive process that is influenced by several external factors including electric and magnetic fields, temperature, pH, chemicals, space limitation and molecular crowding. These factors influence the ability of Proteins To fold into their correct functional forms.
Extreme temperatures affect the stability of Proteins and cause them to unfold or denature. Similarly, extreme pH, mechanical forces and chemical denaturants can denature Proteins. During denaturation, Proteins lose their tertiary and secondary structures and become a random coil. Although denaturation is not always reversible, some Proteins can re-fold under certain conditions [7].
Diseases caused due to protein miss functioning
Proteins can miss function for several reasons. When a protein errors at folding it can cause denaturation of the protein. Denaturation is the loss of protein structure and function [8].
This miss folding does not always result in complete lack of function, but there is a partial loss of function. This miss functioning of Proteins can lead to different diseases in the human body.
Alzheimer’s disease
Alzheimer’s Disease (AD) is a progressive neurological disorder that cause the brain to shrink (Atropy) and brain cells to die. Alzheimer’s disease is the most common cause of dementia and a continuous decline in several mental functions. In America alone 5 million people are affected by it, including nearly half of those who are 85 years or above [9]. The major risk factors of Alzheimer’s disease are age, family history, and heredity. AD typically results in a continuous decline in thinking, behavioral and social skills that affects a person’s ability to function independently, changes in mood [10]. AD results in dense plaques in the brain that are comprised of fibrillar β-amyloid Proteins along with β-sheet secondary structure [11]. In the brain matter, these plaques visually appear to look like voids and connect directly to the decline or deterioration of thought processes. AD has been established as a case of misfolded Proteins the brain, where the misfolded protein is directly related to the formation of plaques in the brain [12].
It is yet to be known what causes the misfolding of the protein initially. However, there was several theories that point out to the oxidative stress in the brain as the triggering stage.
Cystic fibrosis
Cystic Fibrosis (CF) is a hereditary disease that affects the lungs and digestive system of the patient. The body produces thick and sticky mucus that can clog the lungs and obstruct the pancreas which is life-threatening as it prevents proper food processing [13]. CF is a chronic disease that affects 30,000 Americans and less than a million cases per year in India. CF is also a result of protein misfolding. The misfolding leads to some changes in protein which is known as Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) that results in a possible fatal disease [14]. In majorly 70% of CF cases, in the CFTR, the phenylalanine is deleted at position of 508. This depletion of Phe508 is in direct connection of CF formation [15]. The protein misfolding that results in CF occurs prior to birth, but the reason is not entirely known as to why it is caused.
Reasons for protein folding failure
Folding allows a protein to adopt a shape for a specific function. However, this folding is missed due to some reasons such as, when an individual human possess a gene mutation that changes an amino acid in the protein to find its suitable fold. This is generally found in inherited genes, where the mutation influences only a particular protein and its relative function. For example – mutations that lead to cystic fibrosis and sickle cell anemia. In some cases, protein folding failure is an ongoing process and occurs commonly. When Proteins are synthesized, the system that reads the directions from DNA to create long chains of amino acids can make blunders. Scientists estimate that this system, the ribosome, can cause errors in as many as one in every seven Proteins! These errors can result in Proteins that are less likely To fold properly [16]. Even if a protein has no mutation or errors, it still may not fold into its preferred shape or native state. This happens simply because Proteins do not necessarily fold with 100% accuracy. Factors such as acidity and temperature fluctuations in the cell influence the protein folding that differs from one organism to another.
Coping mechanism of cells
Since protein folding occurs frequently, the cells are accustomed to coping with this dysfunction. The cells have several systems in place to refold or destroy aberrant protein formations.
The first in line of defence are the system called Chaperones. Appropriately named, they partner with the Proteins through the entire folding process, thereby improving the chances of a protein To fold suitably and also providing another chance for misfolded Proteins along the way to refold again. Interestingly, chaperones are Proteins themselves. There are various types of chaperones. Some serve specifically in helping a particular type of protein fold, while the remaining act generally. Some chaperones are in the shape of large hollow cubicles and offer the protein a safe space to settle in, isolated from other molecules, where they can fold. The production of the chaperones is boosted in number when a cell is encountered with high temperatures or other extreme conditions thereby making the protein folding a more difficult process, thus earning these chaperones the alias, “heat shock Proteins.”
Another such in line for cell defense against the misfolded Proteins are called Proteasome. If misfolded Proteins wander in the cell, they are most likely to be targeted for destruction and picked up by this machine that chews them up and spits them out as small fragments of amino acids. The proteasome is like a recycling center that allows the reuse of amino acids to produce more Proteins within the cell. Proteins frequently assemble to form larger structures to perform important cellular functions. For example, the tail of a human sperm is a structure that is composed of different types of Proteins that collectively work together to perform a single action, here they form into a network of rotary engine which propels the sperm forward [6].
- Fraser JS, Clarkson MW, Degnan SC, Erion R, Kern D, et al. (2009) "Hidden alternative structures of proline isomerase essential for catalysis". Nature 462: 669–673. Link: https://go.nature.com/3iSda2e
- Bu Z, Callaway DJ (2011) Proteins move! Protein dynamics and long-range allostery in cell signaling. Adv Protein Chem Struct Biol 83: 163–221. Link: https://bit.ly/3qd3XTL
- Frauenfelder H, Sligar SG, Wolynes PG (1991) "The energy landscapes and motions of Proteins". Science 254: 1598–1603. Link: https://bit.ly/3qf3Erv
- Lee A (2005) Proteins are not rigid structures: Protein dynamics, conformational variability, and thermodynamic stability. Link: https://bit.ly/3wWWoTy
- Berg JM, Tymoczko JL, Stryer L (2002) Secondary structure: Polypeptide chains can fold into regular structures such as the alpha helix, the beta sheet, and turns and loops. In Biochemistry (5th ed., section 3.3). New York, NY: W. H. Freeman. Link: https://bit.ly/3zIhRBB
- Protein Folding. Susha Cheriyedath. Link: https://bit.ly/35D0DI4
- Protein Structure. Link: https://bit.ly/3qg9NUA
- Garrett RH, Grisham CM (2010) Biochemistry fourth edition; Brooks/Cole. Australia 160: 93-95. Link: https://bit.ly/2SGscgV
- Alzheimer's Disease. Centers for Disease Control and Prevention. 2010. Link: https://bit.ly/3qcjTWr
- Alzheimer's Association. 2011. Link: https://bit.ly/3iX4SGr
- Glenner GG, Wong CW (1984) Alzheimer's disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem BiophysRes Commun120: 885-890. Link: https://bit.ly/35FDJjk
- Hashimoto M, Rockenstein E, Crews L, Mashliah E (2003) Role of Protein Aggregation in Mitochondrial Dysfunction and Neurodegeneration in Alzheimer’s and Parkinson’s Diseases. Neuromolecular Med4: 21-36. Link: https://bit.ly/2Sdav8g
- Cheung JC, Deber CM (2008) Misfolding of the Cystic Fibrosis Transmembrane Conductance Regulator and Disease. Biochemistry 47: 1465-1473. Link: https://bit.ly/3iTjlDb
- Koppaka V, Axelsen P (2000) Accelerated Accumulation of Amyloid β Proteins on Oxidatively Damaged Lipid Membranes. Biochemistry 39: 10011-10016. Link: https://bit.ly/3j4GXEK
- Riordan JM, Rommens JM, Kerem B, Alon N, Rozmahel R, et al. (1989) Identification of the cysticfibrosisgene: cloning and characterization of complementary DNA. Science 245: 1066-1073. Link: https://bit.ly/3xBpuIb

Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.
 Save to Mendeley
Save to Mendeley
