Open Journal of Bacteriology
Mechanism of multi-resistant bacterial pathogenesis: mdr genes are not so deadly unless plasmid-mediated toxin, virulence and regulatory genes are activated
Kousik Poria, Shampa Bhatta, Sanatan Das, Madhumita Dey, Chandan Halder, Sankalita Datta and Asit Kumar Chakraborty*
Cite this as
Poria K, Bhatta S, Das S, Dey M, Chakraborty AK, et al. (2020) Mechanism of multi-resistant bacterial pathogenesis: mdr genes are not so deadly unless plasmid-mediated toxin, virulence and regulatory genes are activated. Open J Bac 4(1): 008-019. DOI: 10.17352/ojb.000013Mdr genes in association with many drug efflux and metal efflux genes are creating pathogenesis due to antibiotic void. However, most dangerous step occurred when R-plasmids and integrons (~2-9kb) were combined with F’-conjugative plasmid (62.5kb) creating large mdr conjugative plasmids that easily donated 6-15 mdr genes to gut microbiota as well as environmental bacteria. Notably, 2-4x1012 human gut microbiota are very valuable in our body for vitamins synthesis and coenzymes perform >30,000 enzymatic reactions of human and animal metabolosome. It seems mdr gene creation is becoming more easy day by day as plasmids have acquired many gene creation genes like recombinases, DNA polymerases, DNA topoisomerases, integrases and transposases. In truth, antibiotics pressure was so instrumental that 25-40% bacteria of Ganga River and Bay of Bengal water were ampicillin and tetracycline resistant and more than 20 class of β-lactamases and drug transporters were generated in mdr plasmids with millions of mutated isomers increasing drugs MIC. When isolated superbugs were injected into male Wister rats, no detectable toxicity was observed up to 3-6 months follow up. We propose that mdr genes are not so injurious unless toxins and virulence genes with signalling and transcriptional regulators are activated in plasmids. Likely, mdr genes are protected by a tight symbiosis involving bio-film formation in the gut for vitamin synthesis. We also found a gradual increase of larger plasmids in the GenBank database with multiple mdr genes, drug efflux genes, regulatory genes, vitamin synthesizing genes and metal resistant genes as well as abundant (20-40) transposons and IS-elements. WHO, UNDP and CDC have suggested to develop alternate to antibiotics like phage therapy, gene therapy and nanotechnology based toxic drug delivery. We are actively using heterogeneous phyto-antibiotics from Suregada multiflora (root), Cassia fistula (bark) and Trapa bispinosa (fruit peel) targeting superbug central dogma machinery with success.
Introduction
Many toxins like Stx1, RTX, ByB, Enterotoxin, Neurotoxin, Hemolysin, HipA and virulence proteins have been involved in acute bacterial pathogenesis [1,2]. However, recent trends to blame mdr genes (amp, bla, cat, aac, dhfr, str, aph, aad, mcr) and drug efflux genes (tet, mac, acr, mex, mtr) in bacterial pathogenesis have roared due to drug void [3-5]. We blame many toxins and regulatory genes as acute problem in recent bacterial pathogenesis while tremendous amount of transposes and insertion sequences may be serious concern for the alteration and transmission of genes. Now mdr bacteria were detected in river, sea and air as well as in hospital surgical and household matters, claiming 2000,000 deaths per year likely among the neonatal and elderly with weak immune system where 10-20 potent antibiotic therapy were failed [6-9]. So, we re-evaluated the status of mdr genes in plasmids and genome islands in association of toxins and regulatory genes.
Before 1600s we do not know that microorganisms (virus, bacteria, fungus and parasites) cause diseases and we blame ghosts and demons. After the discovery of microscope by Anton Van Leeuwenhoek (1670s) and further pioneering works by Edward Jenner (1790s), Lewis Pasteur (1860s) and David Koch (1880s) proved that bacteria were the culprit of many diseases like TB, Cholera, Gonorrhoea and Typhoid [10]. Eventually the unicellular unseen bacteria were in centre stage of biochemical and molecular biological studies to decipher the central dogma processes like replication, transcription and translation to control diseases by antibiotics. Such study is important as basic chemical reactions of formation of DNA, RNA, protein, sugar, and fat are same among the all life forms. Our body is made of 30-40 trillions cells which are also microscopic (5-100µm). Thus, understanding the molecular assembly of molecular biological processes is vital to design drugs against deadly pathogens [11]. Biomolecules are nanometer and could be analyzed by assembly (107-1015 molecules) using suitable sensitive methods like UV detection of Ethidium bromide stained DNA/RNA, Ninhydrin colour reaction of amino acids, and absorption spectra analysis technologies like MASS, NMR, FT-IR and Raman Spectroscopy [12]. Main point is research is now becoming costly and needs sophisticated machines and talented scientists mostly found in developed nations. Indian peoples are in serious financial crisis as the cost of imported new drugs are very high and almost out of reach for poor patients [13]. Thus we believe in heterogeneous phyto-antibiotics developed by ancient Hindu Civilization.
Discovery of antibiotics that kill bacteria to cure many diseases
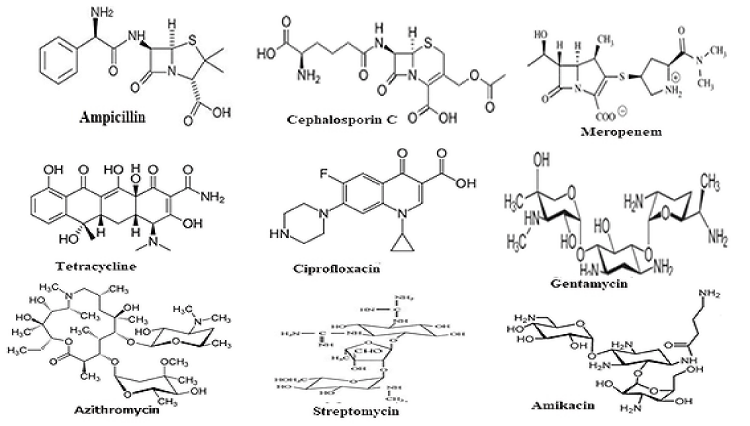
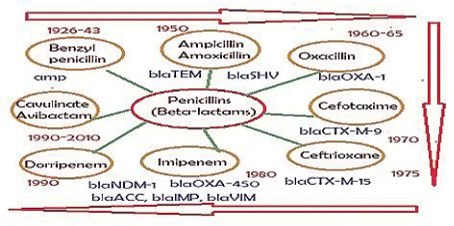
Notably, antibiotics are in centre stage of discoveries since the discovery of penicillin drug by Nobel Laureate Alexander Flaming from slime mold Penicillium notatam in 1928 targeting peptidoglycan cell wall biosynthesis of most Gram(+) and few Gram(-) bacteria [14]. Since then, hundreds derivatives were made alone for penicillins (ampicillin, cefotaxime and imipenem etc) for better drug usually called penicillinases resistant drugs (for chemical structure Figure 1). Dr. Selman Waksman discovered over twenty antibiotics that led to Nobel Prize in Physiology or Medicine in 1952 [15]. Most importantly, streptomycin was alone greatly reduced the TB in 1950s [16]. However, such dream could not last long as more potent penicillinases called, oxacillinases (blaOXA), cefotaximases (blaCTX-M), carbapenemases (blaKPC) were appeared in bacterial plasmids between 1940-1990 increasing drug resistance and antibiotic failure [17-20].
MDR proteins and drug transporters prevent the action of antibiotics and save gut bacteria
Bacteria simply made 100 different enzymes like β-lactamases, acetyl-transferases, phospho-transferases that all could destroy antibiotics by different modes of action [21]. β-lactamases have been grouped into four major classes (A–D) based on sequence homology but diverged more profoundly into 20 sub-classes as described recently [22]. Million of mdr genes have been sequenced that includes mostly bla genes (TEM, SHV, KPC, CTX-M-1/2/9, OXA2/23/48, VIM, CMY, GIM, SPM etc), drug modifying genes like catB3 (chloramphenicol acetyl transferase), aadA1-10 (aminoglycoside adenyl transferase), aacA1-42 (drug acetyl transferase), aph-I-IX (aminoglycoside phosphotransferase), \as well as sul1/2, arr3, dhfr, mcr-1 and vanA genes [23,24]. Diverse range of drug transporter genes like acrAB, macA/B, mexCD/EF, bcr, bmr, tetA/C, norA and mtrCDE are activated in plasmids and chromosome and had arose due to mutation and recombination among 200 ABC transporters that would otherwise regulate the entry and exit of organic and macromolecules into bacterial cells [25]. So, such bacteria in our body stay alive and divide most to cause sepsis and trauma and condition will not be going to improve by taking prescription drugs because no achievable concentration of the drug would be happen in bacterial cytoplasm to stop protein synthesis, RNA synthesis and cell wall synthesis [12]. In truth, drug industry has so far busy to discover new antibiotic derivatives to overcome the actions of mdr genes but now very much reluctant to invest on new antibiotic discovery due to quick drug inactivation creating mdr enzymes [6,7].
mdr conjugative plasmids ubiquitously donate mdr genes into all gut and environmental bacteria
Study confirmed that all early plasmids carried 2-3 mdr genes as in pSC101 or pMB that were used to make semi-synthetic plasmid pBR322 that was sequenced in 1965 to discover the mdr genes amp and tet [18]. It was found that due to plasmid incompatibility (Inc plasmids), one bacterium could not carry multiple different plasmids. However, intake of continuous multiple doses different antibiotics have prevented vitamin synthesis in the gut. So save your soul has worked and bacteria have used its conjugative plasmid (F’) as carrier of mdr genes to transmit genes into new bacteria by a process known as conjugation. More than twenty Tra genes in conjugative plasmids code for proteins that can form sex pili connecting other bacteria to send DNA into it [26-28]. Bacteria combined R-plasmid (3-9kb) with F’-plasmid (62.5kb) and such plasmid was known today as conjugative mdr plasmid which could be large as 50-500kb [29]. Bacteria got very advantage for life as such plasmids are very stable in bacteria during cell division and also can carry multiple mdr genes and can donate the mdr genes to non-mdr bacteria to save from deleterious effects of antibiotics and toxic chemicals [30,31]. In 1928 to 2018, we did lots of chemistry and recombinant DNA technology and gene manipulation but at the end we got PAN drug resistant bacteria! Phage therapy, gene therapy, phyto-antibiotics and nanotechnology are new technologies to avert the drug void. We believe heterogeneous phyto-antibiotics which will rule the future Earth to save human again from superbugs as depicted in ancient Hindu Civilization history like Sanskrit books Charaka Samhita, Sasruta Samhita and Atharva Veda [12].
Toxins and regulatory transcription factors are chromosomal origin
Toxin genes are those that have profound mechanisms to cause severe toxicity in host to create acute pathogenesis. Cholera toxin was discovered in 1959 and contained one subunit A (28KB) and five subunits of B (11KB). While subunit B help to bind membrane GM1 gangliosides, subunit A activates (100x) cytosolic adenyl cyclase by ADP-ribosylation of Gαs (also prevents GDP hydrolysis) to generate cAMP which in turn activates CFTR chloride efflux protein through PKA kinases to exclude Cl- as well as other ions and water to cause watery diarrhoea (2liter/hr) and life threatening dehydration condition [32,33].
Salmonella typhus toxin has three subunits where five PltB subunits make a pyramid-shaped molecule with one molecule each of PltA subunit and CdtB subunit. CdtB has DNase activity while PltA and PltB have ATP ribosylation activity similar to pertussis toxins [34]. Typhoid toxin binds to the podocalyxin-like protein or CD34 sialomucin protein with sialylated glycan (terminal Neu5Gc) in epithelial and endothelial cells or CD45-like protein in hematopoietic cells. It has been postulated that Rab32 GTPase is essential for toxin docking into different human cells to generate immunogenicity and fever. But critically Salmonella toxin can stop cell division at G2/M phase likely due to activation of CDK kinase through P53 TF that induces cyclin inhibitor [35,36]. Bacteria can alleviate toxin toxicity by toxin-antitoxin homeostasis [37-40].
We have analyzed the recent trend of plasmid characteristics in mdr bacteria of Ganga River of Kolkata and propose a model where mdr bacteria are quite safe and such mdr proteins actually protect gut bacteria for vitamin synthesis saving human race. To pinpoint many mdr bacteria were tested in male Wister rats to show minimum effects for long time. A database analysis confirmed the localization of toxin genes, mdr genes, regulatory genes in large plasmids of diverse species of Enterobacteriaceae.
Materials & methods
Water collection and drug sensitivity assay
Water from Ganga River was collected on Monday morning from Babu Ghat (Kolkata-700001). About 100µl of water was spread onto 1.5% Luria Bartoni-agar plate containing different antibiotics at 5-50µg/ml. mdr bacteria were selected in agar-plate containing ampicillin+ streptomycin+ chloramphenicol+ tetracycline or ciprofloxacin at 50, 50, 34, 20 µg/ml respectively. As imipenem and meropenem resistant bacteria were present low (0.08-0.2 cfu/ml water) and a modified method was followed. 2 ml 5x LB media was added into 10 ml Ganga River water or Digha sea water and drug was added at f.c 2-10µg/ml and was incubated 24 hrs to get carbapenem resistant bacterial population [13]. Antibiotics were purchased from HiMenia and stored at 20-50mg/ml at-20OC. Antibiotic papers were also purchased from HiMedia according to CLSI-2017 standard. Antibiotic papers are: Amp-25 (ampicillin), Met-10µg (methicillin), CAZ-30µg (ceftazidime), AT-50 (aztreonam), T-10 (tetracycline), COT-25µg (cotrimoxazole), LOM-15µg (lomofloxacin), VA-10µg (vancomycin), AK-10µg (Amakacin), LZ-10µg (linezolid), TGC-15µg (tigecycline) and IMP-10µg (imipenem).
Isolation of genomic DNA and plasmid DNA
Genomic DNA was isolated by Proteinase-K-SDS method following extraction with phenol-CHCl3-isoamyl alcohol (25:24:1) and ethanol precipitation [9]. 1.5 ml over night culture was spun at 5000rpm and the bacterial pellet was dissolved in 50µl TE buffer (10mM TrisHCl PH 8.0 + 1mM EDTA) and 25µl 10% SDS and 5µl of 20mg/ml proteinase-K were added, incubated for 2-4 hrs and extracted with 25µl 5M sodium chloride and 100µl CHCl3: isoamyl alcohol (25:1). Then centrifuged at 10000 rpm for 10 min. and the DNA pellet was dissolved in 50µl TE buffer, treated 1µl RNase A and extracted with phenol-CHCl3-isomyl alconol (25:24:1) and was precipitated with 1/9 vol. 0f 3M sodium acetate PH 5.2 and 2 volumes of ethanol. The plasmid DNA was isolated from overnight culture using Alkaline-Lysis Method [41]. Simply, to 1.5ml bacterial pellet 100 solution-I was added and mixed well. Then 200µl of cold Solution-II added to make transparent solution and then 150µl cold of Solution-III was added and mixed well and centrifuged at 10000rpm for 10min. To clear solution then added 1 ml 99% ethanol and centrifuged at 10000 rpm for 10 min at 4OC. Plasmid DNAs from four such preparation were combined and the RNA were removed by RNase-A treatment as above and finally plasmid DNA was dissolved in 50µl TE buffer and was stored at -20OC. 0.8% agarose gel electrophoresis in 1x TAE buffer at 50V for 4 hrs was performed to see the plasmid DNAs after staining in 0.5µg/ml ethidium bromide and UV illumination [42].
PCR and DNA sequencing
16S rRNA gene colour Sanger’s di-deoxy sequencing was performed by SciGenom Limited, Kerala, India) [43]. PCR amplification was performed using 1 unit Taq DNA polymerase, 20ng DNA template, 0.25mM dXTPs, 1.5mM MgCl2, for 35 cycles at 95OC/30” (denaturation)-52OC/50”(annealing)-72OC/1.5’ (synthesis). The product was resolved on a 1% agarose gel in 1X TAE buffer at 50V for 4 hrs and visualized under UV light and photograph was taken [12]. mdr genes fragments were purified and were sequenced by di-deoxy method at SciGenom Labs Pvt Ltd, Kerala, India. The primers for 16S rRNA amplification and mdr genes are given below. NCBI BLAST analysis was performed for bacterial specific gene analysis (www.ncbi.nlm.nih.gov/blast) and data was submitted to GenBank.
Preparation of organic phyto-extract (mdr-Cure)
The bark of Suregada multiflora (Ban-Naranga), Cassia fistula (Bandor-Lathi) were collected on July- Novenber 2017-2019 from medium sized tree at Midnapore district of West Bengal. Each 10gms dried and grinded plant and spice parts (Suregada multiflora, Cassia fistula, Syzygium aromaticum, Cinnamomum zeynalicum) was suspended in 40 ml ethanol and overnight extracts were mixed and concentrated 5 times (mdr-Cure) and 50µl used for Kirby-Bauer agar hole assay. Thin Layer Chromatography (TLC) was performed using methanol, water and acetic acid (50:40:10) as mobile phase for 1 hr. Organic molecules were seen and recovered by UV shadowing and was eluted in ethanol from silica-gel. TLC-purified chemicals were purified by HPLC High Performance Liquid Chromatography). Mass spectrometry (Mass), NMR (Nuclear Magnetic Resonance spectrometry) and FTIR (Fourier Transformed Infra Red spectrometry) are then performed to characterize functional groups of active principles in mdr-Cure extract [12].
Animal experiment
Male Wister rats (30-35g) were feed with granules made with (flour 100g + suzi 100g+ mustard oil 100ml+ sugar 100g+one multivitamin tablet). About ~5X106 MDR bacterial suspension (o.5ml) was injected subcutaneously to male rats. Then the rats were challenged with 0.5 ml mdr-Cure extract or 0.5 ml cefotaxime (50mg/ml). After two weeks, blood was collected by tail puncture and bacterial colony forming units were determined on LB-agar plate. The blood smear was drawn on slide and stained with Leishman stain and observed under microscope. Importantly mdr-Cure eliminated 80-90% mdr-bacteria but 3rd generation cephalosporin could not do that. The infected rats were kept for 3-6 months if Escherichia coli KT-1_mdr and Pseudomonas aeruginosa DB-2_mdr could able to kill the rats [30]. But even after 3 months rats were very active demonstrating mdr bacteria were not life threatening unless toxin and virulence genes were activating. The animal experiment was approved by Animal Ethics Committee of OIST.
Result and discussion
R-plasmids and mdr conjugative plasmids with multiple mdr genes
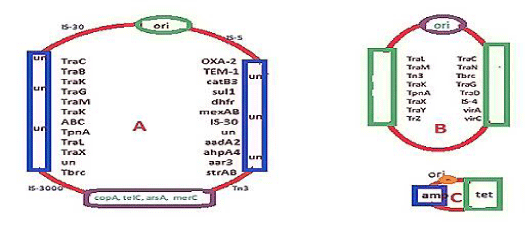
mdr genes were identified in 1965 after the discovery of DNA sequencing and Recombinant DNA Technology. Amp, tet, cat and dhfr genes were identified early between 1940-1965. So small R-plasmids and integrons and IS-elements (transposons) are sequenced in 1965 and onwards as cloning of DNA were introduced and di-deoxy sequencing was available. We see the structure of recombinant pBR322 plasmid where amp and tet genes were located (Figure 2C). In truth pBR322 was a DNA recombinant product of three plasmids preparation from penicillin and tetracycline resistant bacteria, derived by cutting with restriction endonucleases like EcoR1 and HindII, and then joining with DNA ligase enzyme. Such DNA breakage and reunion naturally occurred where 62kb conjugative plasmid (Figure 2B) combined with R-plasmid (Figure 2C) to form super conjugative plasmid (Figure 2A). The primary amino acid sequences of amp and tet mdr genes are given in Figure 3A (AMP beta-lactamase) and Figure 3B (tetracycline efflux protein). We introduced such data to make it clear that such genes were mutated so drastically that hundred of isomers were generated with almost no sequence homology to the parent amp and tet genes. Amp gene was renamed as bla gene. There were at least 20 distinct classes of bla genes like blaTEM, blaCTX-M, blaOXA, blaKPC, and blaNDM etc and each had few hundred isomers so far sequenced from clinical isolates of multidrug-resistant bacteria. Similarly, tet gene has tetC, tetO, tetM, tetS etc isomers and all inactivate tetracycline by drug efflux or drug binding. Notably, huge antibiotic prescription drugs intake killing gut bacteria and preventing vitamin biosynthesis were blamed for the creation of mdr genes but water chemical and metal toxicities were also a concern.
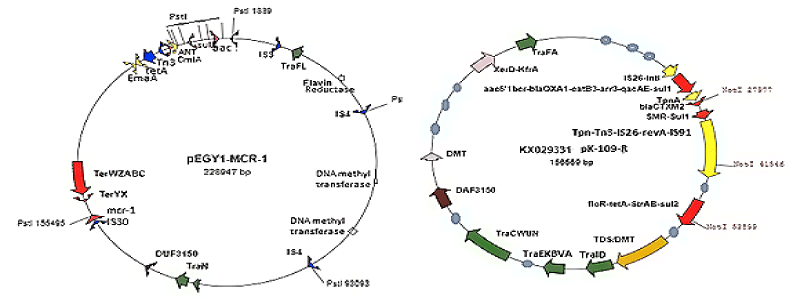
As for now, mdr plasmids are stable and have large space to acquire multiple similar genes needed for new gene creation increasing enzyme concentration. So many mdr plasmids had now multiple integrases (int), recombinases (tnp), DNA topoisomerases (topoIII), reverse transcriptases (maturase) and promoter-enhancers. Since 2010 WHO declared war against mdr bacteria with Action Plan worldwide. Lately, drug companies introduced β-lactamase inhibitor (cavulinate, sulbactam, avibactam) combination therapy but problems are continuing. In truth avibactam still a good drug against penicillinases but not others inhibitors. Most devastating fact, carbapenemsaes are clustered in large conjugative plasmids in presence of ESBL enzymes like KPC-2, VIM, OXA-23, OXA-210 and diverged drug acetyl/phospho transferases including PBPs and also many drug efflux genes (10-15 mdr genes in one plasmid). Those bacteria are named as superbugs and such infections must be treated in the developed countries like UK and USA. A prototype pEGY1 plasmid structure was presented in Figure 4A which had mcr-1 mdr gene (discovered in 2016) that could inactivate colistin drug which so far used in case of superbug infection. In fact drug industry once made new drug derivative to suppress the action of an mdr enzyme, a new beta-lactamase is generated like blaNDM-1 discovered in 2009. The cell wall peptidoglycan biosynthesis is a best target for drug development against bacteria as human has no cell wall, but we fail and bacteria won so far. The generation of diverse types of Beta-lactamases due to action of penicillin, cephalosporin and carbapenem drugs was presented in Figure 5.
K. pneumoniae 151kb plasmid (pKP048) (accession no. FJ628167) contains mdr genes like macrolide ABC transporter, floquinolone resistant gene (qnrB4), mercury resistant gene (MerE), sulphonamide resistant protein (sul1) and aph gene for aminoglycoside phosphotransferases. BlaKPC1 gene was located in K. pneumoniae plasmids (accession nos. NC_022078, NC_014312, JX283456 and KF954759) and blaNDM1 gene was also located in many large conjugative plasmids (CP009116, JN420336 and AP012055) [Dotson et al., 2016]. Plasmid pKOX_R1 contains metal resistant genes and many ABC, MFS, AAA drug transporters as well as common mdr genes like cat, sul1, aac3’-IId, FosA3,ANT, aph and NDM-1, CTX-M-3 and SHV-12 β-lactamases. Most importantly such plasmids has less Tra genes but 16 types of inserted IS-elements and transposons.
Detection of toxin genes in plasmid indicates multi-resistance will create more calamities
Escherichia coli medium sized plasmid (P2; accession no. CP022052.2) has enterohemolysin (protein id. ASE51239; 988aa; RTX Toxin c-terminus) and toxin B (protein id ASE51195; 3169aa) but no mdr genes. Many toxin genes (netB, cpe, cpe2, iota) in plasmids of Clostridium perfringens reported and few are mdr conjugative [37]. The plasmid pCP8533etx (accession no. AB444205; 65kb) has epsilon-toxin and beta-toxin including ABC transporter, thiF vitamin synthesizing gene and DNA methylase but no amp, tet ot strAB ancestral mdr genes were found. The plasmid pCPPB-1 (accession no. AB604032; 67kb) has ioto toxin-ia/ib and enterotoxin including histidine kinase and response regulator genes. The plasmid pCW3 (accession no. DQ366035; 46kb) has acquired tetA and tetM tetracycline resistant genes including DNA methyl transferases and Arc transcriptional regulator but no mdr gene. However, in large plasmid pCLK (266kb), we found neurotoxin cluster (protein ids. ACQ51342, ACQ51274, ACQ51172, ACQ51216) and also hemagglutinin (HA33) RICIN family lectin protein as well as penicillin resistant protein (protein id. ACQ51167; pen1A), AAC acetyl transferase, MATE type drug transporter, DNA methyltransferases, RNA polymerase, DNA polymerase III and ThiH vitamin metabolizing genes. Toxin genes in Clostridium perfringens plasmids (<100kb) were sequenced, As for example, pJIR3537 (netB), pCP8533etx (etx, cpb2), pCPF5603 (cpe, cpb) and pCPPB-1 (cpe, iota) toxin genes but no mdr gene was found (bla, tet, cat, aac, aph or sul1). But small R-plasmid pIP401 has catmdr gene and 47kb pCW3 plasmid has tetM gene for tetracycline binding protein where as no toxin gene has assembled yet.. Similarly, large plasmid pRSJ10_1 (266kb; accession no. CP013684) of Clostridium botulinum has penicillin resistant protein (pen1A), hemagglutinin toxin (protein id. APQ78847), C5 Methyl transferase and vitamin metabolizing genes like thiamine synthase and precorrin methylase. Similarly, in another C. botulinum plasmid pJIR4150 (89kb) has autolysin (protein id. CRG98306), glycosyl transferase toxin (protein id. CRG98303), bacitrin transporter and putative acetyl transferase (accession no. LN835296).
We searched toxin genes in large or small plasmids of Vibrio cholerae. As for example, single mdr gene plasmids are pVCR94deltaX (sul2); pKA1 (dhfr); AY958065 (strA) and multi-mdr genes plasmids are pNDM-116-17 (arr3, cmlA, blaOXA-10, aadA1, mphE, msrE, sul1, blaNDM1); pV1447 (tetD, mel, mph2’, aadA, sul1, dhfr, blaTEM1); and pCR94 (tetC, tetB, catA1, aph3’-1b, aph6’-1d, blaCARB-4, dhfr, QAEdelta, andsul1) were important but no toxin gene indicating STX gene was yet chromosomal (see, accession nos. LN831185, KM083064, NZ_CP033514) (De et al. 1951). We also checked the Salmonells enterica large conjugative plasmids pF8475, p109/9, pHCM1, pB71 and pR64 with 9, 7, 6, 4 and 2 primitive mdr genes (blaTEM, strAB, dhfr, sul1/2, tetB/A, aadA, aph3’, aacC2 etc.) but no toxin genes (accession nos. KP899804, KP899805, AL513383, KP899806, AP005147 respectively). Study indicated that sea, sec, sed,see, eta, and etb enterotoxin genes were associated in Staphylococcus aureus pathogenesis. We detected such genes in many S. aureus plasmids. As for example, 27424bp pSM31 plasmid contained ser,sej, sed enterotoxin genes (protein ids. AKS10460/61/63) and blaZ mdr gene including blaR1 and blaI regulatory genes. Similar plasmid pSA564-fus has enterotoxins (protein ids. ATL64011/12/14) and also in plasmid pSK67 (protein ids. ADA80031/32/34). Similarly, 43396bp pV2200 plasmid contained only sej enterotoxin (protein id. ALK43110) but mph2’ aminoglycoside phosphotransferase, ABC drug efflux protein and blaZ/blaR1/blaI locus involved in induced penicillin resistance. Our hypothesis is if toxin genes are symbiotically protected, then their presence in plasmid would be more random similar to mdr genes. However, the present notion to use gene therapeutics (antisense, ribozyme, siRNA) may alter the genetic signaling (recombination) with rapid acquisition of toxin genes from chromosome into mdr plasmids.
Detection of virulence genes in mdr plasmids during multi-resistant pathogenesis
Virulence genes like fimbriae biogenesis proteins (FimA/B/H), HlyA/D, foeG, estII, iucD, fanA and other regulatory proteins have been implicated in the pathogenesis of Escherichia coli, Salmonella enterica and Mycobacterium tuberculosis. However, such genes were associated with chromosome. But localization of toxin genes, transcription regulatory genes and vitamin synthesis genes in mdr plasmids suggested that vir genes might also be mobilized into mdr conjugative plasmids. In conjugative plasmids pAPEC-02-211A-colV, pAMSC2, and pQnrS1_035148, we found many siderophore genes like aerobactin A/C/D as well as few mdr genes like QnrS1 or dhfr and few drug efflux genes like tetA (protein id. AWX36385) and macB MFS drug efflux gene (accession nos. CP030791, CP031107, CP029368). A FAD-binding protein and cobalamine biosynthesis protein (cobW) were also detected (protein ids. AXF92256, AXF92236). Importantly, Tra genes are deleted in many mdr conjugative plasmids to give place for IS-elements and transposons absolutely required for recombination and transposition to create new mdr gene (blaNDM-16 or mcr-4 or blaOXA-555) against toxic new drug derivatives (imipenem, colistin and levofloxacin etc.).
Mobilization of vitamin metabolizing genes into plasmids to help bio-film symbiosis
We further searched the vitamin metabolizing genes in plasmids to confirm our hypothesis that such genes will be transferred into mdr conjugative plasmids to increase synthesis of vitamins required for >30000 enzymatic reactions in human metabolosome to avert the action of repeated doses of complex antibiotics. In Table 1 we presents data of such GenBank search confirming that thiA/H/I/F/E, cobQ/S,.pdxH, bioT etc genes could be easily detected in plasmids but such genes were very prominent in many Rizobium plasmids. Thus our study confirmed that gut biofilm bacteria still under crisis to continue vitamin synthesis to save intestinal cells.
Mobilization of many transcription factors into mdr conjugative plasmids
GenBank search indicated a overwhelming transposition of transcriptional regulators into mdr plasmids (Table 2). Multi-resistant K. pneumoniae plasmid pKP530 (accession no. KF793937; 61kb) has TetR, LysR and MphR transcriptional regulators at the adjacent of tetA, sul1 and mph2’-1A mdr genes (Table 3). However, such regulators may also control the expression of aac6’-1b-cr (protein id. AHF45947), blaOXA-30 (protein id. AHF45948), blaDHA-1 (protein id. AHF45968) and catB3 (protein id. AHF45949) mdr genes were located in same plasmid likely as poly-mRNA. Salmonella enterica plasmid pA3T (accession no. KX421096; 253kb) fosA3 and OqxB genes were regulated by adjacent tetR and oqxR, and other important mdr genes like blaOXA-1 (protein id. AOR05995), catB3 (protein id. AOR05996), Arr3 (protein id. AOR05997) and ANT3” was also detected in same plasmid. Another SalmonellaTyphimurium mdr strain GDS147 plasmid (accession no. KM877269) contained tnpR regulator at the downstream of dhfr, aadA1/A2 and cmlA chloramphenicol efflux genes. LysR TF (protein id. AKG90162) was also located at the upstream of linked floR, aac6’-1b-cr, sul1, aph, aac3’-IV and blaOXA-1 (protein id. AKG90176) mdr genes. In ESBL plasmid pKP13f (accession no. CP004000; 295kb) located aad3” streptomycin 3” adenyl transferase and aacA4 aminiglycoside 6’ acetyl transferase likely were regulated by transposon promoters; where as dhfr, aadA2, cmlA1, blaCTXM-2, blaOXA-9, blaSHV-12 and blaTEM-1genes might be activated by int or tnpA promoters. A HTH deoR-type transcriptional regulator (protein id. AHE47447; nt. 228311) was located in the middle of such mdr genes. We also propose the roles of tetR (protein id. AFG21523) and LuxR (protein id. AFG21575) TF in Salmonella enterica subsp. enterica serovar Heidelberg plasmid pSH696_135 mdr pathogenesis controlling strAB, sul1, blaAmp-C, blaCMY-2, dhfr, floR and aad2”-IB genes giving multi-resistance (see, accession no. JN983048; Table 3).
pNDM1-010045 plasmid (accession no. CP028560, 190kb) has blaNDM-1 (protein id. AVZ84373), sul2 (protein id. ANZ84383), aac3’-I (protein id. AVZ84355), mph2’ macrolide phosphotransferase (protein id. AVZ84457), and blaOXA-58 ESBL (protein id. AVZ84486). Such mdr genes were likely regulated by ArsR (protein id. AVZ84490), AraC (protein id. AVZ84463), XRE (protein id. AVZ84345) and LysR (protein id. AVZ84385) located in the same plasmid including IS-3, IS-4 and IS-30 elements. Similarly, K. pneumoniae plasmid pPMK1-NDM (accession no. CP008933; 304kb) has three blaNDM1 genes at nt. 47947, 56535 and 65123 including blaOXA1 and blaCTX-M beta-lactamse isomers giving complete resistance to penicillin, cephalosporins and carbapenems. MerR type transcriptional regulator may a important in this plasmid as well as other mdr genes like aacA4, dhfr, aac6’-Ia, and ANT3”-Ia. In K. pneumoniae 154kb plasmid pCR14_2 (accession no. NZ_CP015394) we detected OXA-2 enzyme including mdr genes, aac6’-1b, QacE∆, sul1, CTX-M-2, TEM-1 and aac3’-IIa, clustered at nt. 111745-127396 region and many Tra genes (TraI/K/B/V/W/U/N) and surprisingly nt. 1-40000 region had many unknown genes (DUF domain proteins). Unusually no distinct transcriptional regulaor was detected but an ArsR-like TF.
The plasmids discussed above have no great toxin gene to discuss but the plasmids which had toxins as in Table-3 might have many transcriptional regulators. As for example, Enterobacter cloacae large plasmid p22ES-469 nine transcriptional regulators like LexA, HTH, AcrR, UhpA, FadR, Zur, LysR, AraC, and SoxR (protein ids. PIA01506, PIA01710, PIA01737, PIA01760, PIA01793, PIA015031, PIA01796, PIA01716, and PIA01585 respectively) were accumulated. However, only GCN5 acetyl transferases and MFS drug efflux proteins were detected but no beta-lactamase gene. Sugar metabolising and vitamin synthesis genes were important aspect of this plasmid and likely were activated by numerous transcription factors (Table 3). So such analysis clearly indicated that toxin genes are rare in mdr plasmids but mdr genes are abundant with prominent transcriptional regulators.
Pseudomonas aeruginosa strain AR_0111 XDR clone mdr-island (accession no. CP032257) has many mdr genes (blaOXA-4, ANT3”-Ia, cmlA, tetG, aac3;-Ia, sul1, vanA, mexEF, PBPs) and many transcription factors like LysR (protein ids. AXZ90574, AXZ90543, AXZ95626, AXZ95001, AXZ95625, AX93389, AXZ93271, AXZ94943), HlyD (protein ids. AXZ95115), AraC (protein ids. AXZ95059, AXZ95038, AXZ95648, AXZ94991, AXZ94944, AXZ93336, AXZ95582), MexT (AXZ95056), GntR (AXZ93413), TenA (protein id. AXZ90500) and HTH (protein id. AXZ93247). Such results clearly demonstrated that chromosomal DNA as origin of transcriptional regulators and such genes mobilised into plasmids by recombination. If mdr-islands have role in transferring the vit and tf genes into plasmids is not known. But once it is acquired by conjugative plasmid, then propagation will be occurred by conjugation. That way more and more tf and vit genes will be found in mdr conjugative plasmids in the future.
Gut biofilm and mdr genes are in symbiosis for vitamin synthesis and antibiotic void
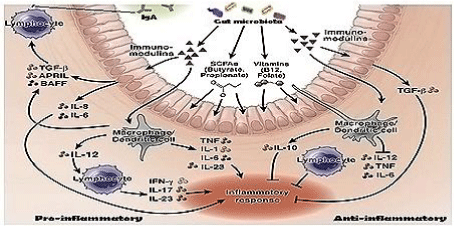
We use 200,000 mt antibiotics per year. We are so depend on antibiotic use that survey has indicated common people like prescription drug even the disease is viral and auto-immunity will be developed within one week. Thus, every man use 10-20 prescription doses of antibiotics per year destroying gut microbiota disrupting the symbiosis between bacteria and intestinal cells. We believe that huge use of antibiotics are cause of multi-resistance today as well as metabolic diseases like diabetes, indigestion, tension and cancer. It is believed that lipopolysaccharides are in a signalling cascade to produce interleukins and cytokines by intestinal cells controlling biofilm (Figure 6).
Thus mdr gene creation is to protect human and animal from extinct although such discrepancies are balanced by prescription drugs like multi-vitamin capsule and probiotic capsule. As the higher derivatives are protective to some extent, amoxillin + cavulinate or cefotaxime, azithromycin and ofloxacin are very much used in India. However, meropenem, amikacin, linezolid are best drugs although blaOXA-58, blaKPC-2, blaNDM-1, aph-III-1b, aac-6’-1b-cr are mdr genes that could be effective in maintaining PAN multi-resistance. We detected the 0.002% bacteria have such genes over 45% bacteria have blaTEM, tet, dhfr, sul, and cat types early mdr genes. BlaNDM1 gene was detected in 2009 and mcr-1 gene was detected in 2016 making imipenem and colistin drugs useless. Scientists detected >500 mutant isomers for each blaTEM, blaOXA, blaCTX-M and blaAmp-C, where as blaOXA has at least 12 distinct sequence types with only 20-40% similarities at specific zones. It appeared that multiple antibiotics use have greatly facilitated the recombination among the various plasmids in mdr bacteria and it was just few days to months to make a new genotype with increase in drug MIC. Such discussion confirmed that (i) we will continue to use antibiotics, (ii) mdr genes will recombine to make more deadly mdr gene isomer (iii) we will face more health hazards, (iv) multi-vitamin and bifidobacteria capsule or curd will improve the antibiotic action (v) more diabetes, mental disorders and cancer will follow if we disturb bio-film and (vi) greedy politicians and rich men will bring World War III to save themselves from distressed people.
Injection of mdr bacteria onto rats have no immediate ill effects
When we tested the mdr-cure extract on rats, we found that 93% mdr bacteria Escherichia coli KT-1_mdr were eliminated by phyto-extract but rats appeared healthy for long time. So we continued the study. We intradermally injected E. coli KT-1_mdr and P. aeruginosa PB-2_mdr five different positions but no death of the animals up to three months and rats were shown very active with weight gain (Figure 7). The mdr bacteria were characterized in Figure 8 demonstrating plasmid profiles, blaTEM and blaCTX-M mdr genes PCR detection in isolated plasmids and inhibition of those mdr bacteria by different phyto-extracts. Growth rate was good and after one month bacterial load could be detected in blood but three experiments showedno death of animals between 3-4 months. Such bacteria were confirmed for blaTEM, blaCTX-M, tetA, acrAB, and aac6’-1b genes (24,30). Thus, we concluded that mdr genes were not so injurious to health but only did create the antibiotic void. However, those bacteria were imipenem sensitive. This data was supported US Microbiome Project where many mdr genes were sequenced from bacteria isolated from normal individual [44.45].
Conclusion
Life creation is due to chemical evolution with assembly of many bio-molecules (amino acids, lipids, nucleotides, carbohydrates and vitamins). Derwin’s adaptation theory tells the unique way of changes in cellular structure and metabolism in different eco-system About 40% of peoples reside at the bank of Ganga River and many big industries are made nearby releasing many pollutants and excretory matters which support lower life forms but also toxicity present. In Ganga River, we see number of bacteria is increasing with 45% resistant to semi-synthetic drugs ampicillin and amoxicillin and 26% resistant to tetracycline. Such drugs had cured most infections between 1940-1990 although gradual increase in drug-resistance pathogens were detected in many continents as early as 1960. Thus, superbugs were highly contaminated in water resources of Ganga River, Bay of Bengal and likely rest of the world. G20 leaders formulated the action plan against mdr bacteria spread. WHO warned that if alternative to antibiotics would not discovered, very fatal human loss might be occurring in the future? High antibiotic concentration in global water is concern due to huge population increase with antibiotic consumption and use of antibiotics in agricultural land and livestock production. Although toxin genes are main regulators of pathogenesis but ¼ unknown genes surely have some roles in bio-film formation and gut microbiota protection and thus maintaining mdr pathogenesis. Further, high rate antibiotic uses in expression vectors containing inducible operon (tetO, blaO) for enzymes and anti-metabolites production have also been questioned [13].
We presented the data that other than mdr genes, many important genes were assembled in mdr Conjugative Plasmids increasing the calamity of multi-resistance. As for example, toxin genes (Table 1), vitamin genes (Table 2), transcriptional regulatory genes (Table 3) were detected in plasmids. The modern plasmid proto-type structure was presented in Figure 4 and mechanism of new β-lactam drugs vs beta-lactamase isomers generation with time was presented in Figure 3. Thus, study of mdr genes to control multi-resistance may not be possible and other plasmid associated genes must be addressed. So our effort to describe the plasmid associated toxin and regulatory genes is important to know the regulatory issues of plasmid-mediated pathogenesis due to augment expression toxins and virulence genes. Likely herbal antibiotics research has given priority in India as there are enough medicinal plants and spices available as described in Sanskrit books Charaka Samhita, Sasruta Samhita and Atharva Veda [46]. New technologies like gene medicines (ribozymes, miRNA, antisencse RNA, and metal nanotechnology) have been welcome to stop the horror of mdr bacterial pathogenesis. We believe in herbal heterogeneous antibiotics and we demand conservation of medicinal plants worldwide. We slogan “save plants and use as medicine” to save millions of peoples those are dragged into nasty poverty line each year due to high cost of antibiotics. Of course, drug void will increase with time as 20-60 transposable elements and rDNA recombinases were assembled in plasmids making easy new mdr gene creation [20,47,48]. In summary, we proved that mdr genes are not immediate threat. We believe that mdr genes will be rescued if we stop huge complex antibiotic intake. That way injectables may be better than oral antibiotics. Anyway assembly of too many mdr genes in plasmids and chromosome islands forced scientists to discover new targets and new methods for the control of bacterial pathogenesis like phage therapy, gene therapy and nanodrug carriers. But immediate high cost of drugs dragged millions of peoples to poverty line as no insurance coverage in the poor African and Asian countries. Thus, drug resistance is a brand name discussed everywhere as in cancer therapy and viral infection and likely a dark age of antibiotics has appeared and thus easy use of antibiotics and anti-cancer drugs may not be a solution today complicating medical field. Surely, drug efflux proteins (AcrAB/CD, MexAB/CD/EF, MacAB, MtrCD) in plasmids are causing more threat to medicine use in clinical therapies.
We thank Late Dr Jagat Bandhu Medda of Oriental Association of Education and Research for funding. AKC is Associate Professor of Biochemistry at OIST. Shampa Bhatta is Ex-student of Biotechnology, Sankalita Datta is MSC 4th Sem Biochemistry student and SD, MD, CH are MSC 4th Sem Biotechnology students, Vidyasagar University. Kousik Poria is IIT GATE qualified PhD student.
Conflict of interest and Ethical Issues
None has involved in any conflict of interest and other ethical issues.
- Blower TR, Short FL, Rao F, Mizuguchi K, Pei XY, et al. (2012) Identification and classification of bacterial Type III toxin-antitoxin systems encoded in chromosomal and plasmid genomes. Nucleic Acids Res 40: 6158-6173. Link: https://bit.ly/2JFhHSR
- Fozo EM, Makarova KS, Shabalina SA, Yutin N, Koonin EV, et al. (2011) Abundance of type toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic Acids Res 38: 3743-3759. Link: https://bit.ly/2UFnq14
- Okeke IN, Laxmaninarayan R, Bhutta ZA, Duse AG, Jenkins P, et al. (2005) Antimicrobial resistance in developing countries. Part 1: recent trends and current status. Lancet Infect Dis 5: 481-493. Link: https://bit.ly/2xML5DU
- Chakraborty AK (2016) Multi-drug resistant genes in bacteria and 21st Century problems associated with antibiotic therapy. Biotechnol Ind J 12: 113-114. Link: https://bit.ly/2xKDXIc
- McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, et al. (2013) The Comprehensive Antibiotic Resistance Database. Antimicrob Agents Chemther. 57: 3348-3357. Link: https://bit.ly/2UFnBtg
- Laxminarayana R (2014) Antibiotic effectiveness: Balancing conservation against innovation. Science 345: 1299-1301. Link: https://bit.ly/2UEgEsG
- Xu ZQ, Flavin MT, Flavin J (2014) Combating multi-drug resistant gram-negative bacterial infection. Exp. Opini. Investi. Drugs 23: 163-182. Link: https://bit.ly/3aCwHg5
- Davis J, Davies D (2010) Origins and Evolution of Antibiotic Resistance. Microbiol Mol Biol Revi 74: 417-433. Link: https://bit.ly/2V31dcf
- Chakraborty AK (2015) High mode contamination of multi-drug resistant bacteria in Kolkata: mechanism of gene activation and remedy by heterogenous phyto-antibiotics. Indian J Biotechnol 14:149-159. Link: https://bit.ly/3aKMVUC
- Tortora GJ, Funke BR, and Case CL (2016) Microbiology: An Introduction. 12th edi. ISBN: 9780321929150. Link: https://amzn.to/2xPUp9X
- Sigeris HE (1987) A History of Medicine: Early Greek, Hindu and Persian Medicine, vol. II, New York, Oxford University Press. ISBN: 9780195050790. Link: https://amzn.to/2UYN7J7
- Chakraborty AK (2017) Ganga action plan, heterogeneous phyto-antibiotics and phage therapy are the best hope for India tackling superbug spread and control. Indian J Biological Sciences 23: 34-51. Link: https://bit.ly/2UGvRt9
- Chakraborty AK, Poira K, Saha D, Halder C, Das S (2018) Multidrug- Resistant Bacteria with activated and diversified MDR Genes in Kolkata Water: Ganga Action Plan and Heterogeneous Phyto-Antibiotics tackling superbug spread in India. American J Drug Delivery Therapeutics 5: 1-9. Link: https://bit.ly/2ypECPx
- Flaming A (1928) On the antibacterial action of cultures of a Penicillium with special reference to their use in the isolation of B. influenzae. British J Exp Pathol 10: 226-236. Link: https://bit.ly/3bS9ab0
- Waksman SA, Woodruff HB (1940) The soil as a source of microorganisms antagonistic to disease-producing bacteria. J Bacteriol 40: 581-600. Link: https://bit.ly/3aFnB2g
- Schatz A, Bugle E, Waksman SA (1944) Streptomycin, a substance exhibiting antibiotic activity against gram-positive and gram-negative bacteria. Exp Biol Med 55: 66-69. Link: https://bit.ly/3c198xZ
- Abraham EP, Chain E (1988) An enzyme from bacteria able to destroy penicillin. Rev Infectious Diseases 1940. 10: 677-678. Link: https://bit.ly/2wSgogJ
- Boliver F, Rodriguer RL, Greene PJ (1977) Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2: 95-113. Link: https://bit.ly/3aGjpz3
- Chakraborty AK (2016) Multi-drug resistant genes in bacteria and 21st Century problems associated with antibiotic therapy. Biotechnology: An Indian Journal. 12: 114 Link: https://bit.ly/2xKDXIc
- Chakraborty AK (2019) Heterogeneous phyto-antibiotics and other future therapeutics against multi-drug resistant bacteria. Advances Biochemistry 7: 34-50. Link: https://bit.ly/3dV1WF6
- Chakraborty AK (2016) Complexity, heterogeneity, 3-D structures and transcriptional activation of multi-drug resistant clinically relevant bacterial beta-lactamases. Trends Biotechnol-open access 2: 1-001.
- Chakraborty AK (2019) Current status and unusual mechanism of multi-resistance in Mycobacterium tuberculosis. J Health Med Informatics 10: 328. Link: https://bit.ly/2UZaHWa
- Kumarasamy K, Kalyanasundaram A (2012) Emergence of Klebsiella pneumoniae isolate co-producing NDM-1 with KPC-2 from India. J Antimicrob Chemother 67: 243-244. Link: https://bit.ly/2UGXfas
- Chakraborty AK, Pradhan S, Das S, Maity M, Sahoo S, et al. (2019) Complexity of OXA Beta-Lactamases involved in Multi-Resistance. British J Bio-Medical Res 3: 772-798.
- Sun J, Deng Z, Yan A (2014) Bacterial multidrug efflux pumps: Mechanisms, physiology and pharmacological exploitation. Biochem Biophys Res Communi 453: 254-267. Link: https://bit.ly/2ysRcNX
- Pansegrau W, Balzer D, Kruft V, Lurz R, Lanka E (1990) In vitro assembly of relaxosomes at the transfer origin of plasmid RP4. Proc Natl Acad Sci. USA 87: 6555-6559. Link: https://bit.ly/3aIvkfU
- Paterson ES, More MI, Pillay G, Cellini C, Woodgate R, et al. (1999) Genetic analysis of themobilization and leading regions of the IncN plasmids pKM101 and pCU1. J Bacteriol 181: 2572-2583. Link: https://bit.ly/2xLHp5u
- Frost LS, Ippen-Ihler K, Skurray RA (1994) An analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol Rev 58: 162-210. Link: https://bit.ly/2yyiOkZ
- Botts RT, Apffel BA, Walters CJ, Davidson KE, Echols RS (2017) Characterization of Four Multidrug Resistance Plasmids Captured from the Sediments of an Urban Coastal Wetland. Front Microbiol 8: 1922. Link: https://bit.ly/2V6zxDI
- Chakraborty AK (2017) Multidrrug resistant bacteria from Kolkata Ganga River with heterogeneous MDR genes have four hallmarks of cancer cells but could be controlled by organic phyto-extracts. Biochem Biotechnol Res 5: 11-23. Link: https://bit.ly/347lVw1
- Guo Q, Spychala CN, McElheny CL, Doi Y (2016) Comparative analysis of an IncR plasmid carrying armA, blaDHAA1 and qnrB4 from Klebsiella pneumoniae ST37 isolates. J Antimicrob Chemother 71: 882-886. Link: https://bit.ly/2V4nDu1
- Ganguly NK, Kaur T (1996) Mechanism of action of cholera toxin & other toxins. Indian J Med Res 104: 28-37. Link: https://bit.ly/2UH90O1
- Spangler BD (1992) Structure and function of cholera toxin and the related Escherichia coli heat-labile enterotoxin. Microbiol Rev 56: 622-647. Link:
- Galán JE (2016) Typhoid toxin provides a window into typhoid fever and the biology of Salmonella Typhi. Proc Natl Acad Sci USA 113: 6338-6344. Link: https://bit.ly/3dXb8ZU
- Dougan G, Baker S (2014) Salmonella enterica serovar Typhi and the pathogenesis of typhoid fever. Annu Rev Microbiol 68: 317–336. Link: https://bit.ly/2xQVkqE
- Song J, Gao X, Galán JE (2013) Structure and function of the Salmonella Typhi chimaeric A(2)B(5) typhoid toxin. Nature 499: 350–354. Link: https://bit.ly/2whzD2U
- Gerdes K, Christensen SK, Lobner-Olesen A (2005) Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol 3: 371-382. Link: https://bit.ly/39AoasM
- Park SJ, Son WS, Lee BJ (2013) Structural overview of toxin-antitoxin systems in infectious bacteria: a target for developing antimicrobial agents. Biochim Biophys Acta 1834: 1155-1167. Link: https://bit.ly/2yyVXWr
- Wang X, Wood TK (2011) Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl Environ Microbiol 77: 5577-5583. Link: https://bit.ly/34djLem
- Wozniak RA, Waldor MK (2009) A toxin-antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet. 5: e1000439. Link: https://bit.ly/2JCbJlx
- Ausubel FM, Brent R (1989) In Current Protocols in Molecular Biology. Greene publishing associates and Wiley-Interscience, New York.
- Maniatis T, Fritsch EF, Sambrook, J (1982) Molecular Cloning-A Laboratory Manual. Cold Spring Harbour Laboratory, 1982. ISBN: 978-0879691363. Link: https://amzn.to/2X6Ruo4
- Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA 74: 5463-5467. Link: https://bit.ly/2JzFQdz
- Le Chatelier E (2013) Richness of human gut microbiome correlates with metabolic markers. Nature 500: 541-546. Link: https://go.nature.com/2X4tIZX
- Jandhyala SM (2010) Role of the normal gut microbiota. World J Gastroenterol 21: 8787-8803. Link: https://bit.ly/2V05jlB
- Chakraborty AK (2018) Nucleic-Acids Based Nanocarriers, in “Nanocarriers for Drug Delivery”. eds. Mahapatra et al. chapter-5; pp.155-172, Elsevier Press, Amsterdam. ISBN:978012814.640.
- Chakraborty AK (2020) Chemical toxicities of both Intestine and environmental water caused genetic recombination in bacteria with the creation of MDR genes and drug void. EC Pharmacology and Toxicology 8: 1-14.
- Chakraborty AK (2018) Poor correlation of diversified MDR genes in Gonococci plasmids: Does alteration in chromosomal DEGs, PBP2 and target Mutations sufficient to wide spread multi-resistance in Neisseria gonorrhoeae? J Health Med Informat 9: 1-8. Link: https://bit.ly/39AoF68

Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.



 Save to Mendeley
Save to Mendeley
