Journal of Surgery and Surgical Research
The differential usage of molecular machinery in brain cancer patients with iron-enriched glioma environments
Brandon Lucke-Wold1*, Michael Joseph Diaz1, Joanna Song2, Sai Batchu3, Kevin Root1, Karan Patel4 and Kamil Taneja5
2University of South Florida, Morsani College of Medicine, USA
3Montville, NJ, United States, USA
4Rowan University, Cooper Medical School, USA
5Stony Brook University, Renaissance School of Medicine, USA
Cite this as
Lucke-Wold B, Diaz MJ, Song J, Batchu S, Root K, et al. (2022) The differential usage of molecular machinery in brain cancer patients with iron-enriched glioma environments. J Surg Surgical Res 8(3): 030-035. DOI: 10.17352/2455-2968.000150Copyright
© 2022 Lucke-Wold B, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Introduction
Gliomas are neuroepithelial tumors in the brain or spinal cord that arise from glial or precursor cells and include astrocytomas, oligodendrogliomas, and ependymomas. They are the most common malignant primary central nervous system tumors, representing 75% of cases in adults and 24% of all cases of primary brain and CNS tumors [1,2]. Despite radiotherapy and temozolomide chemotherapy, which are the current gold-standard first-line treatments, patients with gliomas, particularly the highly aggressive and most common subtype glioblastoma multiforme, still have a poor prognosis with a median survival time of 15 months and 5-year survival rates of 5% [3-6]. Increasing evidence has demonstrated the potential of immunotherapies, including immune checkpoint inhibitors and antigen-specific cancer vaccines, for treating glioma [4-7]. However, these treatments demand a comprehensive understanding of the biological pathways involved in the glioma disease course. Bioinformatics has contributed significantly to the discovery of prognostic markers for gliomas [8], such as the IDH1/2-mutation [9] and MGMT methylation [10], thereby representing a promising approach toward elucidating the genes, pathways, and mechanisms associated with human gliomas.
In trace amounts, iron is an essential element used for basic cellular functions and growth and is used by numerous cell types, including neurons and glial cells where it is a cofactor in enzymes involved with synthesizing and metabolizing neurotransmitters, myelinating neurons, transporting oxygen, and producing ATP through oxidative phosphorylation [11]. When present unbounded and excessively in the body, iron has also been indicated as a risk factor for many diseases, which also include cancers [12,13]. Glial cells are at particular risk of iron overload and thus these pathologies since they are the cells with the highest concentration of iron in the brain [14]. As a cofactor, iron often alternates between its oxidation states, ferrous (Fe2+) and ferric (Fe3+) iron. High amounts of these redox reactions in iron overload can cause free radical formation, such as hydroxyl radicals through the Fenton reaction, and subsequent DNA and protein damage as well as lipid peroxidation, all of which can ultimately result in tumorigenesis or ferroptosis [15]. Tumor cells exhibit iron-sequestering, through upregulating iron uptake and downregulating iron export pathways as well as diverting immune cells located in the tumor microenvironment (TME), which is necessary to support the increased ATP production for cell proliferation and to induce the expression of genes involved in the epithelial to mesenchymal transition (EMT) in these cells during cancer [16,17].
Besides transformation into tumors, cells in the presence of excess iron can undergo cell death due to iron-mediated oxidative damage, a process known as ferroptosis. These dying cells then release damage-associated molecular patterns that activate the immune response and can either stimulate anti-tumor immunity to suppress tumorigenesis or induce an inflammatory response that promotes tumor growth in the TME [18]. Because of its role in tumorigenesis, ferroptosis has been the target of several chemotherapies, such as hepatocellular carcinoma [19], pancreatic cancer [20], colorectal cancer [21] and breast cancer [22]. Changes in the expression of genes involved in ferroptosis have also been associated with numerous other cancers [23], including gastric [24], ovarian [25] and lung cancer [26]. Regarding gliomas and other neurological tumors, ferroptosis and other iron metabolism are less studied, despite current knowledge of how excessive iron plays a role in cancer. However, the presence of databases, such as FerrDb [27] which contains known regulators and markers of ferroptosis and ferroptosis-disease associations, facilitates the use of high-throughput bioinformatic analyses to better understand how high levels of iron in glioma TME affect cancer progression from a cellular perspective.
Traditional exploration of the cellular pathways and sets of genes implicated in cancer activity commonly recruits gene set enrichment analysis (GSEA) [28], which requires sequencing data from two groups (e.g., case and control) to calculate a logarithmic fold change in gene expression and create a ranked gene list. This means that GSEA cannot be applied to studies that only measure subjects of one class, such as The Cancer Genome Atlas (TCGA) (https://www.cancer.gov/tcga) which contains many large genomic studies of only cancer patients. To solve this issue, the non-parametric, unsupervised method of Gene Set Variation Analysis (GSVA) [29] can be used instead of GSEA for similar functional analyses. GSVA calculates gene set enrichment scores in an analogous approach to conventional gene set tests but computes variation of gene set enrichment in samples independent of class labels, bypassing the need for explicit case/control classes.
The current study addresses the sparsity of research into high iron TME in brain cancers by studying expression changes of the gene in iron-metabolism pathways in glioma patients and assessing whether iron enrichment correlates with survival distinctions. Since our patient cohort is only glioma patients, we also employ GSVA in the study, which has not been used for studies on similar topics.
Methods
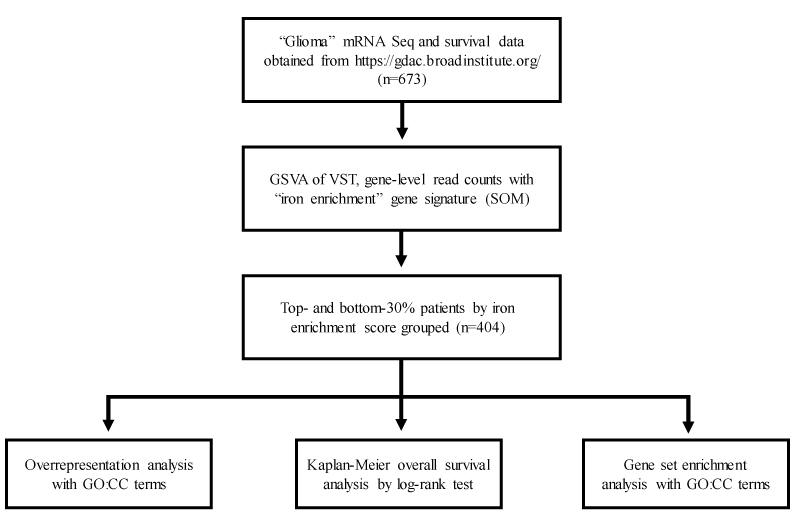
Figure 1 for a detailed schematic of the study design.
Data acquisition: Level 3 mRNA-Seq data and corresponding representing six hundred seventy-three (673) glioma patients with available overall survival information were obtained from the Broad Institute GDAC Firehose portal (https://gdac.broadinstitute.org/). Rounded, gene-level raw counts calculated by RNA-Seq by Expectation-Maximization were used for iron enrichment scoring. Analyzed specimens were obtained post-surgically (without treatment) and with treatment. Radiation therapy-level differences were not expected to appreciably influence total cellular mRNA expression. Table 1 for relevant tumor sample metadata.
Estimating iron enrichment: Tumor-microenvironment (TME) iron enrichment scores were estimated by gene set variation analysis (GSVA) using a merged gene signature constituent of previously published ferroptosis drivers and markers from the FerrDB web tool (http://www.zhounan.org/ferrdb/)[27]. GSVA was run with variance-stabilized counts using the R package ‘GSVA v1.44.2.
Case stratification: Enrichment scores for patients with >1 tumor sample were averaged. Case IDs among the top 30% of iron enrichment scores (ES: 0.05 to 0.35) represented the iron-enriched group (n = 202); case IDs among the bottom 30% of iron enrichment scores (ES: -0.32 to -0.1) represented the iron-depleted group (n = 202). A top and bottom 30% cutoff was applied to restrict downstream analyses to only differentially enriched (or non-zero ES) tumor samples.
Statistical analysis: All statistical analyses were conducted using R v1.4.3. Kaplan-Meier (KM) curves comparing the abovementioned case ID groups were generated using the R package ‘survminer’ v0.4.9. Iron-enriched case IDs were subjected to overrepresentation (log2 fold change (l2fc) > 1.5, min) and gene set enrichment analysis (l2fc > |1.5|) using gene ontology cellular component (GOCC) terms with R package ‘cluster profile’ v4.4.4. Input differentially expressed genes were pre-filtered by adjusted p- value < 0.05 and above described l2fc cutoffs after fold change shrinkage. GOCC terms with reported Q-values < 0.01 were visualized using the R package ‘enrichplot’ v1.16.1.
Results
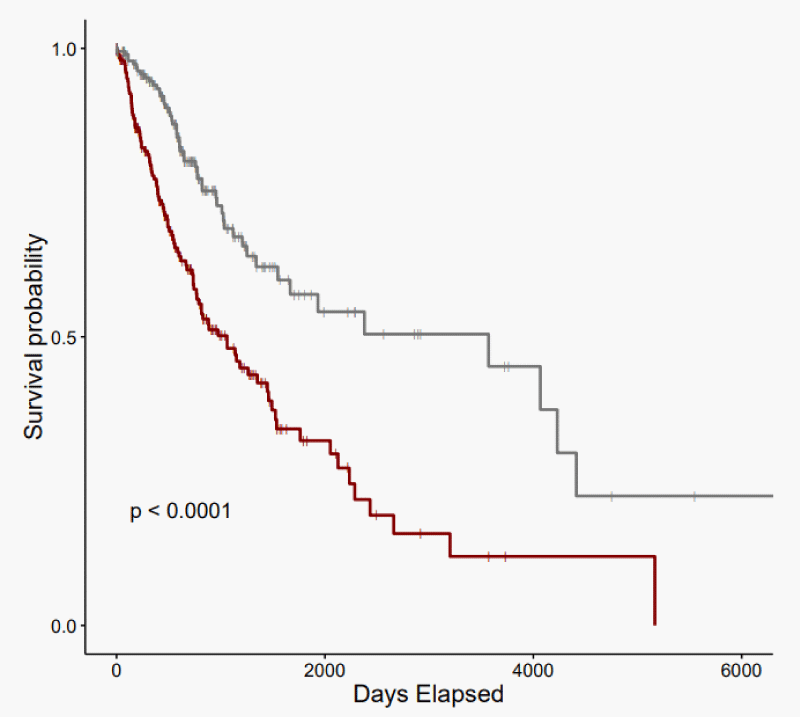
KM analysis revealed that glioma patients with an iron-enriched TME reported worse overall survival (OS) rates than patients with an iron-depleted TME (p - value = 6.1e-07) (Figure 2). The iron-enriched subgroup observed a median OS of 1062 days (95% CI: 771 - 1458). The iron-depleted subgroup observed a median OS of 3571 days (95% CI: 1666-NA).
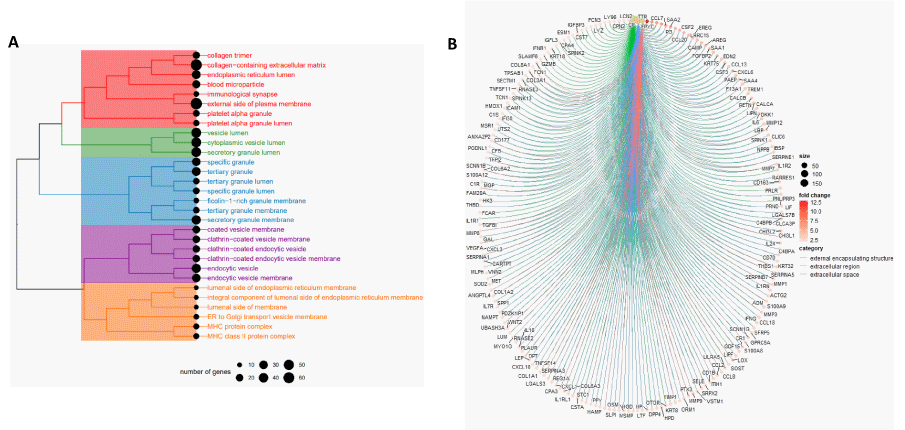
To resolve the predominating cellular components in iron-enriched glioma, overrepresentation analysis (ORA) was performed using the set of differentially expressed genes that are upregulated in the iron-enriched tumor population (Figure 3A). Of the top 30 most significant overrepresented GOCC terms, five distinct clusters were identified: 1) connective tissue and extracellular matrix components; 2) non-specific vesicle lumen components; 3) neutrophil components; 4) endocytosis components; 5) membrane-associated components not described by remaining clusters.
Gene set enrichment analysis (GSEA) was then employed with GOCC terms to functionally annotate the global set of differentially expressed genes among the iron-enriched population (Figure 3B). The top-3 most significant GOCC terms were “external encapsulating structure,” “extracellular region,” and “extracellular space.” These GO sets all represented positive normalized enrichment scores, indicating strong gene product localization to the extracellular domain in iron-enriched gliomas, compared to the group of iron-depleted gliomas.
Discussion
Here we obtained RNA-sequencing data from 673 glioma patients and calculated an iron enrichment score using GSVA based on the expression of established ferroptosis marker and driver genes from FerrDB to identify “iron-enriched” and “iron-depleted” subgroups. Representative case IDs were then subjected to Kaplan-Meier analysis to analyze survival differences. We reported that patients with ferroptosis-promoting glioma reported worse survival compared to those with low expression of similar genes (p - value < 0.0001, Figure 1). These results corroborate with other studies [30,31] which indicate that ferroptosis-related gene sets have prognostic value for human gliomas based on models created using different patient datasets and risk scores calculated via different computational methods (e.g., LASSO regression, random survival forest). Besides these differences, our study utilized a more comprehensive set of ferroptosis-related genes and suggests that increased ferroptosis is negatively correlated with patient outcomes likely as it is an indicator of high levels of iron in the TME.
Moreover, our study also investigated gene set enrichment in these iron-enriched individuals using Gene Ontology (GO) enrichment analyses of Cellular Component terms (CC). GOCC results categorized the genes into five clusters that can be summarized as a cluster pertaining to connective tissue and Extracellular Matrix (ECM) components, a cluster related to non-specific vesicle lumen components, a cluster consisting of neutrophil components, a cluster composed of endocytosis components and a cluster of membrane-associated components not in other clusters (Figure 2A). The cluster pertaining to connective tissue and ECM components derive from the presence of collagen-related, blood and platelet GOCC terms which are either component of the ECM or connective tissue [32]. The cluster consisting of neutrophil components was named because specific granules, tertiary granules and ficolin-1-rich granules are characteristic of neutrophils and these terms made up the majority of GOCCs found in this cluster [33,34]. The cluster composed of endocytosis components was defined because many GOCCs either contained terms explicitly denoting endocytosis or clathrin, a molecule that plays a critical role in one form of receptor-mediated endocytosis [35]. The cluster composed of non-specific vesicle lumen components summarized GOCCs that contained the term “vesicle lumen”, or for the case of secretory granule, is a vesicle, but that did not fit in other clusters that also include GOCCs with “vesicle lumen” in its term. The presence of these last two clusters is likely explained in part by high amounts of enzymes involved in iron uptake and metabolism within a cell, which is done by clathrin-mediated endocytosis [36], which naturally accompanies an iron-enriched TME. Finally, GOCCs that did not seem to fit with the other clusters but contain the term “membrane” or “MHC”, which are found on all nucleated cell membranes in the case of major histocompatibility complex class I or on all antigen-presenting cell membranes in the case of major histocompatibility complex class II, were thus summarized as a cluster of membrane-associated components, not in other clusters.
Within the connective tissue and extracellular matrix components cluster, there is high expression of collagen synthesizing genes such as COL1A1, COL1A2, COL3A1, COL6A2, COL6A3, and COL8A1 as well as LOX, which encodes an enzyme, lysyl oxidase, involved in collagen production, that all display at least a 2 times log fold-change in expression (Figure 2b). Previous evidence demonstrates that collagen acts as a scaffold to guide glioma cell migration in vitro [37,38] and is associated with an angiogenic shift and faster tumor growth and invasion [39,40]. Collagen has also been suggested to be a prognostic marker as more organized collagen architecture is correlated with less invasive glioblastoma xenografts and longer patient survival [41]. While not shown in cells of the central nervous system, excess iron has been shown to increase collagen production in rat hepatic stellate cells [42]. Along with collagen, overexpression of canonical markers of angiogenesis, including vascular endothelial growth factor A (VEGFA), Wnt Family Member 2 (WNT2) and various matrix metallopeptidases (MMP), namely MMP3, MMP7, MMP8, MMP9, and MMP12, indicates extensive ECM remodeling in the gliomas of iron-enriched patients. Interestingly, one gene with relatively high log fold-change, epiphycan (EPYC), is also known to influence ECM organization and has been demonstrated to predict worse prognoses in ovarian cancer [43]. Similarly, transthyretin (TTR), another gene with relatively high log fold-change has been implicated in angiogenesis-related to pathologies of the brain [44] and other parts of the body [45,46]. These studies along with our results suggest that the worse survival outcomes in these patients could be due to excess iron contributing to increased collagen production and ECM remodeling for greater angiogenesis.
Finally, the cluster consisting of neutrophil components points to evidence of an increased inflammatory response in the glioma TME of iron-enriched patients as inflammation has been shown to recruit neutrophils in cancer and non-cancer situations [47,48]. This is further supported by increased expression of inflammation-related genes, including interferon-gamma (INFG) and IL6 interleukin-6 (IL6) (Figure 2B). Neutrophil-induced ferroptosis is also linked to increased necrosis in glioblastoma and poorer survival outcomes, theorized possibly because signals released from ferroptosed tumor cells that worsen neuroinflammation. The precipitated inflammation can then cause a cerebral cytokine storm that results in irreversible damage, organ dysfunction and death [49]. A large number of immune-related genes with increased expression in the iron-enriched patients in our study compared to the iron-depleted patients (i.e., high log fold-change) reinforces this hypothesis and could explain another factor that contributes to the poorer survival outcomes in these patients.
However, as our results are purely based on in silico analyses using public databases, it is imperative to validate our computational analysis through experimental and clinical research. Nevertheless, our study can guide future investigations of how high iron environments can affect neurological tumors, particularly regarding potential biological pathways and genes on which to focus.
Conclusion
This present study reveals that glioma TME enriched for iron correlates with worse GBM survival outcomes. We identified glioma patients with an iron-enriched and an iron-depleted tumor environment based on the combined expression of various ferroptosis markers and drivers and then examined the groups for survival distinctions, of which the iron-enriched group reported poorer OS rates. We further studied the differential expression of genes between the groups and leveraged GO term enrichment to analyze subcellular differences. This analysis provides evidence of higher levels of a neutrophil-driven response, inflammation, angiogenesis and ECM remodeling which may all play a role in explaining the worse prognoses of iron-enriched glioma patients.
- Davis ME. Epidemiology and Overview of Gliomas. Semin Oncol Nurs. 2018 Dec;34(5):420-429. doi: 10.1016/j.soncn.2018.10.001. Epub 2018 Nov 2. PMID: 30392758.
- Lapointe S, Perry A, Butowski NA. Primary brain tumours in adults. Lancet. 2018 Aug 4;392(10145):432-446. doi: 10.1016/S0140-6736(18)30990-5. Epub 2018 Jul 27. PMID: 30060998.
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005 Mar 10;352(10):987-96. doi: 10.1056/NEJMoa043330. PMID: 15758009.
- Huang B, Zhang H, Gu L, Ye B, Jian Z, Stary C, Xiong X. Advances in Immunotherapy for Glioblastoma Multiforme. J Immunol Res. 2017;2017:3597613. doi: 10.1155/2017/3597613. Epub 2017 Feb 19. PMID: 28299344; PMCID: PMC5337363.
- Thakkar JP, Dolecek TA, Horbinski C, Ostrom QT, Lightner DD, Barnholtz-Sloan JS, Villano JL. Epidemiologic and molecular prognostic review of glioblastoma. Cancer Epidemiol Biomarkers Prev. 2014 Oct;23(10):1985-96. doi: 10.1158/1055-9965.EPI-14-0275. Epub 2014 Jul 22. PMID: 25053711; PMCID: PMC4185005.
- Delgado-López PD, Corrales-García EM. Survival in glioblastoma: a review on the impact of treatment modalities. Clin Transl Oncol. 2016 Nov;18(11):1062-1071. doi: 10.1007/s12094-016-1497-x. Epub 2016 Mar 10. PMID: 26960561.
- Huang J, Liu F, Liu Z, Tang H, Wu H, Gong Q, Chen J. Immune Checkpoint in Glioblastoma: Promising and Challenging. Front Pharmacol. 2017 May 9;8:242. doi: 10.3389/fphar.2017.00242. PMID: 28536525; PMCID: PMC5422441.
- Chao B, Jiang F, Bai H, Meng P, Wang L, Wang F. Predicting the prognosis of glioma by pyroptosis-related signature. J Cell Mol Med. 2022 Jan;26(1):133-143. doi: 10.1111/jcmm.17061. Epub 2021 Nov 23. PMID: 34816605; PMCID: PMC8742236.
- Wick W, Hartmann C, Engel C, Stoffels M, Felsberg J, Stockhammer F, Sabel MC, Koeppen S, Ketter R, Meyermann R, Rapp M, Meisner C, Kortmann RD, Pietsch T, Wiestler OD, Ernemann U, Bamberg M, Reifenberger G, von Deimling A, Weller M. NOA-04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with procarbazine, lomustine, and vincristine or temozolomide. J Clin Oncol. 2009 Dec 10;27(35):5874-80. doi: 10.1200/JCO.2009.23.6497. Epub 2009 Nov 9. Erratum in: J Clin Oncol. 2010 Feb 1;28(4):708. PMID: 19901110.
- Weller M, Stupp R, Hegi ME, van den Bent M, Tonn JC, Sanson M, Wick W, Reifenberger G. Personalized care in neuro-oncology coming of age: why we need MGMT and 1p/19q testing for malignant glioma patients in clinical practice. Neuro Oncol. 2012 Sep;14 Suppl 4(Suppl 4):iv100-8. doi: 10.1093/neuonc/nos206. PMID: 23095825; PMCID: PMC3480248.
- Ward RJ, Zucca FA, Duyn JH, Crichton RR, Zecca L. The role of iron in brain ageing and neurodegenerative disorders. Lancet Neurol. 2014 Oct;13(10):1045-60. doi: 10.1016/S1474-4422(14)70117-6. PMID: 25231526; PMCID: PMC5672917.
- Fonseca-Nunes A, Jakszyn P, Agudo A. Iron and cancer risk--a systematic review and meta-analysis of the epidemiological evidence. Cancer Epidemiol Biomarkers Prev. 2014 Jan;23(1):12-31. doi: 10.1158/1055-9965.EPI-13-0733. Epub 2013 Nov 15. PMID: 24243555.
- Torti SV, Manz DH, Paul BT, Blanchette-Farra N, Torti FM. Iron and Cancer. Annu Rev Nutr. 2018 Aug 21;38:97-125. doi: 10.1146/annurev-nutr-082117-051732. PMID: 30130469; PMCID: PMC8118195.
- Reinert A, Morawski M, Seeger J, Arendt T, Reinert T. Iron concentrations in neurons and glial cells with estimates on ferritin concentrations. BMC Neurosci. 2019 May 29;20(1):25. doi: 10.1186/s12868-019-0507-7. PMID: 31142282; PMCID: PMC6542065.
- Wang Y, Yu L, Ding J, Chen Y. Iron Metabolism in Cancer. Int J Mol Sci. 2018 Dec 27;20(1):95. doi: 10.3390/ijms20010095. PMID: 30591630; PMCID: PMC6337236.
- Sacco A, Battaglia AM, Botta C, Aversa I, Mancuso S, Costanzo F, Biamonte F. Iron Metabolism in the Tumor Microenvironment-Implications for Anti-Cancer Immune Response. Cells. 2021 Feb 2;10(2):303. doi: 10.3390/cells10020303. PMID: 33540645; PMCID: PMC7913036.
- Müller S, Sindikubwabo F, Cañeque T, Lafon A, Versini A, Lombard B, Loew D, Wu TD, Ginestier C, Charafe-Jauffret E, Durand A, Vallot C, Baulande S, Servant N, Rodriguez R. CD44 regulates epigenetic plasticity by mediating iron endocytosis. Nat Chem. 2020 Oct;12(10):929-938. doi: 10.1038/s41557-020-0513-5. Epub 2020 Aug 3. PMID: 32747755; PMCID: PMC7612580.
- Chen X, Kang R, Kroemer G, Tang D. Broadening horizons: the role of ferroptosis in cancer. Nat Rev Clin Oncol. 2021 May;18(5):280-296. doi: 10.1038/s41571-020-00462-0. Epub 2021 Jan 29. PMID: 33514910.
- Louandre C, Marcq I, Bouhlal H, Lachaier E, Godin C, Saidak Z, François C, Chatelain D, Debuysscher V, Barbare JC, Chauffert B, Galmiche A. The retinoblastoma (Rb) protein regulates ferroptosis induced by sorafenib in human hepatocellular carcinoma cells. Cancer Lett. 2015 Jan 28;356(2 Pt B):971-7. doi: 10.1016/j.canlet.2014.11.014. Epub 2014 Nov 12. PMID: 25444922.
- Yamaguchi Y, Kasukabe T, Kumakura S. Piperlongumine rapidly induces the death of human pancreatic cancer cells mainly through the induction of ferroptosis. Int J Oncol. 2018 Mar;52(3):1011-1022. doi: 10.3892/ijo.2018.4259. Epub 2018 Jan 31. PMID: 29393418.
- Guo J, Xu B, Han Q, Zhou H, Xia Y, Gong C, Dai X, Li Z, Wu G. Ferroptosis: A Novel Anti-tumor Action for Cisplatin. Cancer Res Treat. 2018 Apr;50(2):445-460. doi: 10.4143/crt.2016.572. Epub 2017 May 10. PMID: 28494534; PMCID: PMC5912137.
- Ma S, Henson ES, Chen Y, Gibson SB. Ferroptosis is induced following siramesine and lapatinib treatment of breast cancer cells. Cell Death Dis. 2016 Jul 21;7(7):e2307. doi: 10.1038/cddis.2016.208. PMID: 27441659; PMCID: PMC4973350.
- Li J, Cao F, Yin HL, Huang ZJ, Lin ZT, Mao N, Sun B, Wang G. Ferroptosis: past, present and future. Cell Death Dis. 2020 Feb 3;11(2):88. doi: 10.1038/s41419-020-2298-2. PMID: 32015325; PMCID: PMC6997353.
- Hao S, Yu J, He W, Huang Q, Zhao Y, Liang B, Zhang S, Wen Z, Dong S, Rao J, Liao W, Shi M. Cysteine Dioxygenase 1 Mediates Erastin-Induced Ferroptosis in Human Gastric Cancer Cells. Neoplasia. 2017 Dec;19(12):1022-1032. doi: 10.1016/j.neo.2017.10.005. Epub 2017 Nov 13. PMID: 29144989; PMCID: PMC5686465.
- Basuli D, Tesfay L, Deng Z, Paul B, Yamamoto Y, Ning G, Xian W, McKeon F, Lynch M, Crum CP, Hegde P, Brewer M, Wang X, Miller LD, Dyment N, Torti FM, Torti SV. Iron addiction: a novel therapeutic target in ovarian cancer. Oncogene. 2017 Jul 20;36(29):4089-4099. doi: 10.1038/onc.2017.11. Epub 2017 Mar 20. PMID: 28319068; PMCID: PMC5540148.
- Alvarez SW, Sviderskiy VO, Terzi EM, Papagiannakopoulos T, Moreira AL, Adams S, Sabatini DM, Birsoy K, Possemato R. NFS1 undergoes positive selection in lung tumours and protects cells from ferroptosis. Nature. 2017 Nov 30;551(7682):639-643. doi: 10.1038/nature24637. Epub 2017 Nov 22. Erratum in: Nature. 2022 Sep 14;: PMID: 29168506; PMCID: PMC5808442.
- Zhou N, Bao J. FerrDb: a manually curated resource for regulators and markers of ferroptosis and ferroptosis-disease associations. Database (Oxford). 2020 Jan 1;2020:baaa021. doi: 10.1093/database/baaa021. PMID: 32219413; PMCID: PMC7100629.
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005 Oct 25;102(43):15545-50. doi: 10.1073/pnas.0506580102. Epub 2005 Sep 30. PMID: 16199517; PMCID: PMC1239896.
- Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013 Jan 16;14:7. doi: 10.1186/1471-2105-14-7. PMID: 23323831; PMCID: PMC3618321.
- Zhuo S, Chen Z, Yang Y, Zhang J, Tang J, Yang K. Clinical and Biological Significances of a Ferroptosis-Related Gene Signature in Glioma. Front Oncol. 2020 Nov 20;10:590861. doi: 10.3389/fonc.2020.590861. PMID: 33330074; PMCID: PMC7718027.
- Wan RJ, Peng W, Xia QX, Zhou HH, Mao XY. Ferroptosis-related gene signature predicts prognosis and immunotherapy in glioma. CNS Neurosci Ther. 2021 Aug;27(8):973-986. doi: 10.1111/cns.13654. Epub 2021 May 10. PMID: 33969928; PMCID: PMC8265949.
- Halper J, Kjaer M. Basic components of connective tissues and extracellular matrix: elastin, fibrillin, fibulins, fibrinogen, fibronectin, laminin, tenascins and thrombospondins. Adv Exp Med Biol. 2014;802:31-47. doi: 10.1007/978-94-007-7893-1_3. PMID: 24443019.
- Segal AW. How neutrophils kill microbes. Annu Rev Immunol. 2005;23:197-223. doi: 10.1146/annurev.immunol.23.021704.115653. PMID: 15771570; PMCID: PMC2092448.
- Rørvig S, Honore C, Larsson LI, Ohlsson S, Pedersen CC, Jacobsen LC, Cowland JB, Garred P, Borregaard N. Ficolin-1 is present in a highly mobilizable subset of human neutrophil granules and associates with the cell surface after stimulation with fMLP. J Leukoc Biol. 2009 Dec;86(6):1439-49. doi: 10.1189/jlb.1008606. Epub 2009 Sep 9. PMID: 19741154.
- Kumari S, Mg S, Mayor S. Endocytosis unplugged: multiple ways to enter the cell. Cell Res. 2010 Mar;20(3):256-75. doi: 10.1038/cr.2010.19. Epub 2010 Feb 2. PMID: 20125123; PMCID: PMC7091825.
- Mayle KM, Le AM, Kamei DT. The intracellular trafficking pathway of transferrin. Biochim Biophys Acta. 2012 Mar;1820(3):264-81. doi: 10.1016/j.bbagen.2011.09.009. Epub 2011 Sep 22. PMID: 21968002; PMCID: PMC3288267.
- Rubenstein BM, Kaufman LJ. The role of extracellular matrix in glioma invasion: a cellular Potts model approach. Biophys J. 2008 Dec 15;95(12):5661-80. doi: 10.1529/biophysj.108.140624. Epub 2008 Oct 3. PMID: 18835895; PMCID: PMC2599859.
- Yang YL, Motte S, Kaufman LJ. Pore size variable type I collagen gels and their interaction with glioma cells. Biomaterials. 2010 Jul;31(21):5678-88. doi: 10.1016/j.biomaterials.2010.03.039. PMID: 20430434.
- Noreen R, Chien CC, Chen HH, Bobroff V, Moenner M, Javerzat S, Hwu Y, Petibois C. FTIR spectro-imaging of collagen scaffold formation during glioma tumor development. Anal Bioanal Chem. 2013 Nov;405(27):8729-36. doi: 10.1007/s00216-013-7337-8. Epub 2013 Sep 26. PMID: 24068168.
- Huijbers IJ, Iravani M, Popov S, Robertson D, Al-Sarraj S, Jones C, Isacke CM. A role for fibrillar collagen deposition and the collagen internalization receptor endo180 in glioma invasion. PLoS One. 2010 Mar 22;5(3):e9808. doi: 10.1371/journal.pone.0009808. PMID: 20339555; PMCID: PMC2842440.
- Pointer KB, Clark PA, Schroeder AB, Salamat MS, Eliceiri KW, Kuo JS. Association of collagen architecture with glioblastoma patient survival. J Neurosurg. 2017 Jun;126(6):1812-1821. doi: 10.3171/2016.6.JNS152797. Epub 2016 Sep 2. PMID: 27588592; PMCID: PMC5386834.
- Gardi C, Arezzini B, Fortino V, Comporti M. Effect of free iron on collagen synthesis, cell proliferation and MMP-2 expression in rat hepatic stellate cells. Biochem Pharmacol. 2002 Oct 1;64(7):1139-45. doi: 10.1016/s0006-2952(02)01257-1. PMID: 12234617.
- Deng L, Wang D, Chen S, Hu W, Zhang R. Epiphycan Predicts Poor Outcomes and Promotes Metastasis in Ovarian Cancer. Front Oncol. 2021 Nov 23;11:653782. doi: 10.3389/fonc.2021.653782. PMID: 34888227; PMCID: PMC8650094.
- Gião T, Saavedra J, Vieira JR, Pinto MT, Arsequell G, Cardoso I. Neuroprotection in early stages of Alzheimer's disease is promoted by transthyretin angiogenic properties. Alzheimers Res Ther. 2021 Aug 24;13(1):143. doi: 10.1186/s13195-021-00883-8. PMID: 34429155; PMCID: PMC8385857.
- Shao J, Yin Y, Yin X, Ji L, Xin Y, Zou J, Yao Y. Transthyretin Exerts Pro-Apoptotic Effects in Human Retinal Microvascular Endothelial Cells Through a GRP78-Dependent Pathway in Diabetic Retinopathy. Cell Physiol Biochem. 2017;43(2):788-800. doi: 10.1159/000481562. Epub 2017 Sep 27. PMID: 28950253.
- Lee CC, Ding X, Zhao T, Wu L, Perkins S, Du H, Yan C. Transthyretin Stimulates Tumor Growth through Regulation of Tumor, Immune, and Endothelial Cells. J Immunol. 2019 Feb 1;202(3):991-1002. doi: 10.4049/jimmunol.1800736. Epub 2018 Dec 19. PMID: 30567728; PMCID: PMC6344285.
- Blaisdell A, Crequer A, Columbus D, Daikoku T, Mittal K, Dey SK, Erlebacher A. Neutrophils Oppose Uterine Epithelial Carcinogenesis via Debridement of Hypoxic Tumor Cells. Cancer Cell. 2015 Dec 14;28(6):785-799. doi: 10.1016/j.ccell.2015.11.005. PMID: 26678340; PMCID: PMC4698345.
- Huebener P, Pradere JP, Hernandez C, Gwak GY, Caviglia JM, Mu X, Loike JD, Schwabe RF. The HMGB1/RAGE axis triggers neutrophil-mediated injury amplification following necrosis. J Clin Invest. 2015 Feb;125(2):539-50. doi: 10.1172/JCI76887. Epub 2014 Dec 22. Erratum in: J Clin Invest. 2019 Mar 4;130:1802. PMID: 25562324; PMCID: PMC4319429.
- Yee PP, Wei Y, Kim SY, Lu T, Chih SY, Lawson C, Tang M, Liu Z, Anderson B, Thamburaj K, Young MM, Aregawi DG, Glantz MJ, Zacharia BE, Specht CS, Wang HG, Li W. Neutrophil-induced ferroptosis promotes tumor necrosis in glioblastoma progression. Nat Commun. 2020 Oct 27;11(1):5424. doi: 10.1038/s41467-020-19193-y. PMID: 33110073; PMCID: PMC7591536.
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
