Journal of Neurology, Neurological Science and Disorders
Better neural images by combining ultrahigh field strength MRI with innovative MRI sequences
Anuhya Dayal1#, Andin Ngwa2#, Brian Rutt3, Arutselvan Natarajan4* and Edwin Chang4*
2College of Letters and Sciences, University of California Berkeley, Berkeley, California, USA
3Department of Radiology, Radiological Sciences Laboratory, Stanford University, Stanford, California, USA
4Department of Radiology, Molecular Imaging Program at Stanford, Stanford, California, USA
#Both Authors contributed equally to the manuscript
Arutselvan Natarajan, Department of Radiology, Molecular Imaging Program at Stanford, 3155 Porter Drive, Rm. 1106, Palo Alto, CA, 94304, USA, E-mail: anatarajan@stanford.edu
Cite this as
Dayal A, Ngwa A, Rutt B, Natarajan A Chang E (2022) Better neural images by combining ultrahigh field strength MRI with innovative MRI sequences. J Neurol Neurol Sci Disord 8(1): 007-017. DOI: 10.17352/jnnsd.000047Copyright
© 2022 Dayal A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Better MRI scanning technologies and protocols can provide insights into neurological disorders. In this review, we describe the basic concepts of MRI and, in the process, we convey to the reader the relevance of MRI as a high-resolution imaging modality of tissue structure and metabolism. We outline the main parameters for improving MRI resolution and sensitivity for the ultimate goal of optimizing the diagnosis of neurological diseases. A key to obtaining high-resolution images by MRI is the strength of the magnet’s external field strength (Bo). The higher the field strength, the better the signal-to-noise (SNR) of acquired signals. Hence, this results in improved sensitivity and resolution of the reconstructed images. This article recapitulates the advancement of MRI technology towards Ultra-High-Field Strength (UHF) apparatus and the consequent benefits in SNR. Other keys towards improving MRI images include deftly modifying the parameters of longitudinal magnetization relaxation time (T1), transverse magnetization relaxation time (T2), repetition times between radiofrequency (RF) pulses (TR), and the time of reading post-pulse (TE). Such parameters can be controlled through acquisition software associated with the MRI machines. The review profiles the cumulative efforts by researchers to complement UHF-MRI hardware with innovative RF pulse sequences protocols such as Diffusion Weighted Imaging (DWI), Pulse Gradient Spin Echo (PGSE), Oscillating Gradient Spin Echo (OGSE), Blood Oxygen Level Dependent (BOLD)-MRI and Arterial Spin Label (ASL)-MRI. Collectively, these advances in both MRI hardware and software have pushed the field to attain a mesoscopic level of resolution. Further enhancements in analyzing MRI images through Artificial Intelligence (AI) algorithms may advance resolutions beyond the mesoscopic stage and perhaps even toward the microscopic resolution of living tissue.
Introduction
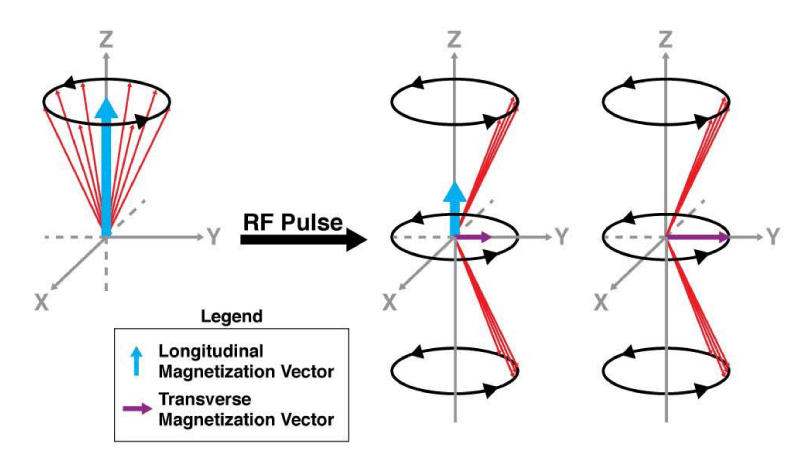
MRI, or magnetic resonance imaging, is an imaging modality that utilizes both magnetic fields and exogenously applied radiofrequency (RF) radiation to create high-resolution visualizations of tissues. Biological tissue is composed largely of water molecules which, in turn, possess a subpopulation of charged hydrogen (known as 1H) protons. When these charged protons are subjected to an external magnetic field (Bo; Figure 1), an intrinsic and charged spin angular momentum is revealed which causes the protons to collectively align and process or quantum mechanically rotate along the axis of the external magnetic field, Bo. The frequency of this precession is known as the Larmor frequency. It is constant and is a function of the type of atom as well as the strength of the external magnetic field. Thus, a hydrogen proton has a different Larmor frequency than say a carbon atom. In addition, the stronger the Bo, the stronger the precession. The precession of the charged atoms causes a magnetic moment to be produced parallel to the axis of B0 and this is known as longitudinal magnetization. At this point, an RF pulse, orthogonal to Bo, can be introduced into the tissue at the Larmor frequency (or the frequency of precession), and this energy is absorbed by the proton to push their precession angle away from the direction of Bo (note that higher Bo results in higher signal above noise). The more perpendicular magnetization is known as transverse magnetization [1] (Figure 2). RF pulses can be strong enough to send the proton to 90o and in some cases 180o (i.e. anti-parallel) to the plane of the external magnetic field, Bo.
Once the RF pulse stops, the precessing protons will not stay in their phased rotation around the Bo axis forever. Environmental inhomogeneities inevitably cause the precessing protons to de-synchronize with one another (a process known as dephasing) and force a realignment along Bo. In the process, energy is released in the form of electromagnetic wave functions. These wave functions are complex with seemingly non-regular periodicity and amplitudes. They are themselves composed of the summation of myriad regular wavefunctions spanning the sampled space. The complex wavefunction can be deconstructed into individual sine or cosine wave components via mathematical operations known as 2D or 3D Fourier transformation. In contrast to the parent wave function, these individual components have a regular wavelength, periodicity, amplitude, and defined phase shift. Such information can be mapped onto an imaging grid called k-space. K-space is conceptually complex and it is beyond the scope of this review to expound upon it in detail. It can best be described as a graphical algorithm that manages to condense the phase pattern and directionality of the Fourier-transformed wave components as data points onto a Cartesian graph. These graphs possess axes (kx and ky) that represent the frequency domains along the x and y-direction of the field of view. The further away from the origin, the greater the frequency. Simply put, the position (kx, ky) of the data points gives information on the frequency and directionality of the individual wave function while the intensity of the data points gives information on the amplitude and phase shift of the wave function. Such information in k-space can be reconstructed into a familiar MRI image through inverse Fourier transformation of all data points with subsequent summation of the wave functions. Further descriptions of this technique (along with the mathematics) are described in more detail elsewhere [2–6].
T1 refers to the constant time required for the longitudinal magnetization to restore after an RF pulse is sent to the tissue. T2 refers to the constant time taken for the transverse magnetization to decay after an RF pulse is sent into the tissue. These two parameters are determined by the type of tissue being imaged, the concentration of water and other molecules, and the parameters of the imaging pulse sequence. These parameters, which are controlled by the experimenter to create the best image, are TR and TE. TR, or repetition time, is the time between RF pulses, or excitations. TE is the time interval after an RF pulse in which the signal emitted from a proton is read. These times of TE and TR can be adjusted to maximize resolution and contrast in an image. Needless to say, both longitudinal and transverse magnetization is accompanied by electromagnetic signals and the signal-to-noise ratio depends upon Bo thus external field strength can also contribute to image resolution.
Images can be T1 or T2 dense, meaning they rely on one of these signals primarily for the signal contrast that produces the image. If an image is both T1 and T2 dense, the result is referred to as a spin dense image, namely, an image that simply reflects the concentration of protons in the tissue. If the image is neither T1 nor T2 dependent, the source of the signal is uncertain, hence the image is not reflective of the tissue being imaged [1]. The image is thus likely an artefact.
These basic concepts create the fundamentals of MR imaging, a current and powerful imaging technology that has great potential to yield information beyond detailed anatomy. How could this be achieved? With variations in pulse sequence, contrast dyes, and imaging techniques, more information than just images can be extracted from these tissues. With advancement allowing imaging at higher field strengths, the question of whether higher field strength can enable enhance imaging is posed. In this review, the advantages and disadvantages of high and ultrahigh field (UHF) strength are explored, and the question of whether mesoscopic imaging is possible today with UHF MRI is investigated.
Materials and methods
This project was completed as a comprehensive review of the existing literature on MRI at various field strengths and the diverse range of imaging techniques that high-field strength MRI enables. The search occurred primarily from the NIH’s National Library of Medicine and PubMed. The keywords used in the first part of our studies focused on the comparison of various field strengths in MRI. Thus, we utilized terms such as “ultrahigh-field strength”, “MRI”, “comparison” and “advantages.” The choice of keywords for the second part of the study focused on techniques and applications. When researching MRI techniques, typical examples were: “diffusion-weighted imaging,” “arterial spin labeling,” “blood oxygen level-dependent,” “AI,” “machine learning,” and “magnetic resonance spectroscopy,”.
Results
As field strength increases, the signal recorded increases which allow for higher resolution (Findings summarized in Table 1) [7].
Here are some more differences to consider in MRI field strengths Table 2:
The images shown in Figures 3a and 3b were taken from a study comparing the image quality of various MRI field strengths in cardiac tissues. The difference in resolution clearly shows increased vessel sharpness of and around the right coronary artery, as well as more detailed vascularization. Quantitatively, the signal-to-noise ratio is higher, as well as the contrast to noise ratio (even though it doesn’t show visually, it is numerically higher) [13].
It also shows the ventricles of the heart at 1.5T, 3T, and 7T. Here, visually the improvement in resolution can be seen; however, quantitatively, the radiomics (numerical data obtained from the image) is about the same, showing no statistically significant difference. While this study doesn’t explicitly ask this question, the question of which is the best field strength to use arises. Field strength alone cannot greatly improve the quality of an image. Instead, optimizing the pulse sequence at that field strength is vital to produce the clearest image possible. Software and hardware work hand in hand in the improvement of resolution in MRI [13].
Currently, one of the strongest MRI machines used in preclinical and clinical studies is at a field strength of 11.7 T. A study that utilized 11.7 T MRI to image ischemic changes in the cerebral cortex of mice after intravoxel incoherent motion. The use of such a powerful scanner allowed the researchers to detect very subtle, but significant, changes in blood flow [14]. The French Alternative Energies and Atomic Energy Commission (CEA) has recently developed an 11.7 T MRI machine with an inner diameter of 90 cm to allow the passage of the whole human body. The conception of a machine this powerful yet still accommodating a large-enough bore to fit a human body is groundbreaking. Most recently, the machine was used to image a pumpkin, to test the machine, and try to optimize the imaging sequences being used. An image with a resolution of 400 microns can currently be achieved, but it is predicted that a machine of this strength should be able to image down to 200 or even 100 microns. Herein lies the immense importance of imaging sequences. While a stronger machine will achieve a higher resolution, researchers first need to figure out how to optimize it. This is usually done using a combination of software and hardware. This year, the CEA was able to achieve the first step towards the creation of a multi-subject probabilistic atlas of the brainstem by using an 11.7T pre-clinical MRI scanner to analyze an ex vivo human brain stem. Because this tissue was obtained post-mortem, their methods involved combining the 11.7 T MRI scanner with various imaging protocols such as Diffusion Weighted Imaging (DWI) to account for the fact that the diffusion coefficient in dead tissues is 4 to 5 times lower than in living tissues. Another factor that was taken into consideration was the T2 relaxation time because specimen fixation does affect the T2 constant, thus the image had to be heavily T2 weighted. Once an image was acquired, different software such as Freeview was used to manually define and segment the acquired 800 slices and WIKIBrainstem Interface was used for real-time 3D navigation of the segmented results. Through this combination of software and hardware, they were able to acquire highly detailed images where small nuclei such as the trochlear nucleus were detected. However, the trade-off with the higher resolution was a loss of contrast which sometimes made it difficult to distinguish between neighboring nuclei [15]. This further exemplifies the importance of optimizing both one’s pulse sequences as well as one’s hardware. Maximizing this potential and developing a methodology on how to image using this 11.7T scanner remains the primary goal of the European Aroma 2022 Project [16]. Optimizing acquisition protocols through the optimization in the sequence and timing of RF pulsing protocols has been a focus of intensive research. The most relevant findings are surveyed in the succeeding section.
MRI imaging techniques
Higher field strength MRI and the consequent higher signal-to-noise ratio allow researchers to observe more information within a scanned image. This is due partly to the increase in sensitivity of high-field strength MRI. But to truly acquire better images one must also rely upon software, in other words, the development of novel and innovative acquisition sequences. These techniques are summarized in Table 3a and explained in further detail below [17].
Spin Echo (SE)
A single RF pulse creates a free induction decay (FID) but the decay is oftentimes so rapid that little useful information can be acquired into k-space. Thus, image reconstruction is impossible. However, two consecutive RF pulses create a spin echo (SE). The spin echo (SE) results in a 180-degree pulse that refocuses dephased protons. The SE regenerates the spin phase information that was lost during the FID. Because these protons process at different speeds due to differences in local microscopic fields, some move faster than others and so these protons lose their phase coherence. An initial 90-degree pulse causes the protons to be transversely magnetized and some of them gain their phase faster than others. When these protons begin to de-phase, their transversal magnetization begins to disappear causing a signal loss. As such, after half of the echo time (TE) has passed, a 180-degree pulse is sent through these protons causing them to process in the opposite directions. Since these protons process at different speeds, the slower-moving protons will now be ahead of the faster-moving ones. After the second half of TE has passed, the faster-moving protons should have caught up with slower ones leading to both a stronger magnetization and signal. This occurs at the center of the spin echo. Beyond this center, the protons begin to de-phase once more. This spin echo pulse sequence only serves as a temporary fix to the constant inhomogeneities of the external magnetic field which affects image quality [18].
Diffusion Weighted Imaging (DWI)
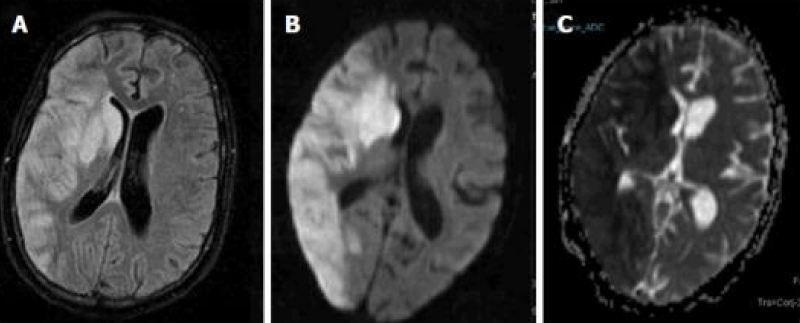
There are, however, variations in the spin echo technique. Diffusion Weighted Imaging (DWI) is a specialized magnetic resonance imaging technique that uses the diffusion of water molecules as a tool for visualizing internal physiology. DWI gives us the ability to non-invasively map out the diffusion processes of water molecules in different biological tissues in-vivo [19]. The diffusion of water molecules in our tissues isn’t random. It follows a pattern based on the intrinsic water properties of various tissue types. Different pathological conditions can cause this pattern to be disrupted leading to a change in the amount of diffusion in the affected area. DWI helps detect these abnormalities, as depicted in Figure 4a. More specifically, DWI is inherently a series of T2 weighted sequences that helps detect the movement of water molecules by applying opposite gradient pulses in each of three orthogonal directions. No net water molecule movement corresponds to the preservation of underlying T2 signal intensity leading to a hyperintense DWI signal. Additionally, a net movement of water molecules along the gradient directions leads to the protons being de-phased and a loss of the underlying T2 signal. DWI signal is higher in retracted diffusion but because DWI contains T2 weighting, areas within the brain with inherently high T2 signal intensities can also show increased signals on DWI sequences so the Apparent Diffusion Coefficient (ADC) is used to clarify whether this is the case. The ADC is a map of values of normal T2 ranges for specific areas of the brain that help distinguish between true diffusion and “T2 shine-through” or areas that have inherently high T2 signals. DWI is especially useful in tumor characterization as well as the detection of acute ischemic infarcts with the key measurement parameter being ADC [20].
Pulsed Gradient Spin Echo (PGSE)
Pulse Gradient Spin Echo (PGSE) is a type of pulse sequence used in Diffusion Weighted Imaging. It aims to provide information about the average diffusion and displacement profiles of particles in a sample [21]. A diffusion sensitizing gradient (DG) is applied to one side of the 180-degree pulse. The phase of stationary spins is unaffected because any phase accumulation will be wiped out by the second DG that will be sent through. Diffusing spins move to different locations at different times between the first and second DG that are sent through. This causes them to fall in and out of phase leading to a signal loss. This difference in pulse sequence is depicted in Figure 4b. Two images are acquired and combined to create an Apparent Diffusion Coefficient (ADC) map. The first image is T2 weighted and is done with the DG turned off or set to a low value and the second image is a Diffusion-weighted sourced image sensitive to diffusion in multiple directions and is done with the DG turned on at various different strengths. The ADC map is used to clarify any abnormalities found in Trace images [22]. PGSE is very important in different applications used to infer cell density, size, and cellular structure [23].
Oscillating Gradient Spin Echo (OGSE)
Oscillating Gradient Spin Echo (OGSE) is a type of pulse sequence utilized in DWI or diffusion-weighted imaging. DWI, as mentioned above, measures the diffusion ability of water molecules in the body to detect anatomical structures. OGSE employs co-sinusoidal-like gradient waveforms that are able to determine diffusion restrictions in its gradient wave [24]. As the frequency of the wave increases, diffusion gets easier, and more measurable parameters are detected at higher scales, allowing for the production of a higher-resolution image (Figure 4b). The use of this technique reveals spatial dimension information that isn’t normally measurable with conventional MR imaging techniques/sequences. For instance, OGSE allows for the measuring of specific neural microstructures such as detecting axon diameter and surface-to-volume ratios. This data can be measured in healthy individuals as well as in patients with various pathological issues to make a comparison regarding neural degradation [25].
Arterial Spin Labelling (ASL)
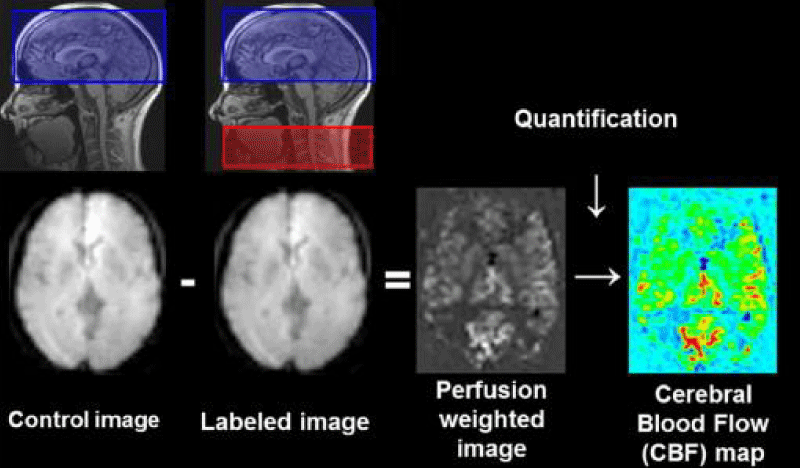
Arterial Spin Labelling (ASL) is a magnetic resonance imaging technique used for measuring tissue perfusion. It uses magnetically labeled arterial blood water protons as an endogenous diffusible intrinsic tracer. ASL provides maps of regional perfusion and does not need any exogenous contrast which is beneficial when dealing with the pediatric population and those suffering from renal insufficiencies. ASL signal-to-noise ratio is inherently low, the signal from the labeled inflowing blood is only 0.5%-1.5% of the full tissue signal [26]. UHF magnetic field strength combined with ASL is beneficial because it increases image SNR and lengthens T1 allowing more spin labels to accumulate. ASL’s main goal is to produce a “flow labeled or tag image” and a “control image” where the static tissue signals are identical but the magnetization of the inflowing blood is different so that when the labeled image is subtracted from the control image, the static tissue signal will be eliminated and the remaining signal will be a relative measure of perfusion proportional to cerebral blood which is the measure of blood circulating in the brain and associate structures (CBF); it helps us determine areas of increased spatial brain activity [26]. Multiple acquired imaged pairs are used to construct a CBF map (Figure 4c). ASL can be used to study the focal hemodynamic pathology of cortical lesions in Multiple Sclerosis [27], differentiate between low-grade tumors and high-grade tumors, and aid in the evaluation of tumor response after treatment [26].
Blood Oxygen Level Dependent (BOLD)
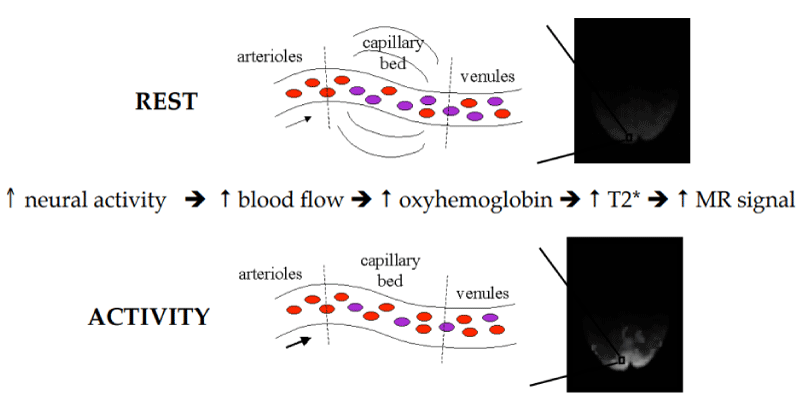
Blood Oxygen Level Dependent MRI (BOLD MRI) is a functional MRI (fMRI) that is used to image metabolic activity in the tissues imaged. As the name suggests, this imaging technique is able to pick up on the slightest variations in blood flow. Hemoglobin, the protein that carries oxygen from the lungs to the tissues of the body, requires iron to be functional. This iron in hemoglobin has an intrinsic magnetic field that changes the magnetic moments of the proximal water molecule protons. This change is then reflected once that tissue is put in an MR machine and subjected to a large magnetic field. Oxygenated hemoglobin is diamagnetic, giving off a higher MR signal, whereas deoxygenated hemoglobin is paramagnetic, giving off a lower MR signal. Hence, the contrast in a TR-dense fMRI image is obtained. Areas of higher signal, or lighter color, indicate more oxygenated hemoglobin, also indicating higher activity/metabolism in those areas (Figure 4di) [27].
BOLD MRI is a vital technique in that it uses endogenous contrast, a method of differentiating between tissues internally rather than through an external dye/contrast agent (i.e. gadolinium). It has been used in neuroscience studies to study cortical activity that correlates with regions of the brain associated with tasks or sensing. It also has been used to assess cerebral microcirculation (blood flow) in detail, and how this changes with various neurodegenerative diseases (Figure 4dii) [17]. Additionally, it has been used to study the role of hypoxia in various bodily organs. Specifically, BOLD has been utilized to assess intrarenal oxygenation levels in patients with chronic kidney disease, kidney injury, renal allograft rejection, and other conditions [29]. Finally, BOLD has also had prospects in fetal imaging. Preventative imaging for the fetus is a field still in need of much research and study, and BOLD MRI can help in this area. Imaging can hypothetically show the risk of hypoxia for the fetus, the maternal oxygenation levels, as well as the perfusion levels of the placenta. Studies in fetal imaging have been limited to animal subjects, but there are promising hopes that the results can be translated for human imaging as well [30].
Discussion
Resolution limits on current UHF MRI: Future perspectives
MR Imaging is a vital and indispensable tool in a physician’s arsenal used to diagnose all kinds of physical and physiological conditions. MR Imaging holds immense potential for furthering medical diagnostic capabilities, treatment tracking abilities, and prognosis evaluation/predictions.
This study serves the purpose of giving a simple and holistic overview of MRI and a wide variety of current techniques and future initiatives to the population of medical and Ph.D. students. The review of the literature aims to provide more insight into the prospects of microscopic imaging utilizing MR technology, diving into and explaining the functionality and applications of spin echo, DWI, ASL, BOLD, AI, MRS, and more.
The benefits that come with ultra-high field MRI such as increased SNR, better spatial resolution, motion correction of movement artifacts, and more, have compelled us to probe if microscopic imaging was possible. After an extensive review of the literature, the results show that although microscopic imaging is not a possibility at this stage, the research field is headed in the right direction. At the moment, mesoscopic resolution and fine anatomy (anatomical details of the tissues but not on a cellular level) are possible at UHF MRI in combination with different techniques such as DWI, ASL, and BOLD. These techniques enable fine detail that can be used to infer cell size, measure tissue perfusion and determine changes in the magnetic field due to blood oxygen levels. One of the current limitations faced is the software that is used to run the MRI machine. Software is just as important as field strength. Since UHF MRIs such as 11.7T MRIs are still new and relatively rare, research has mostly been towards finding radiofrequency pulse sequences that give the best possible image. Other impediments to pushing UHF MRI towards microscopic resolution include non-uniform radiofrequency fields, enhanced susceptibility artifacts, and higher radiofrequency energy deposition in the tissue [31]. Fortunately, novel techniques, such as those mentioned below, are being developed and studied to combat these issues.
AI, Machine Learning (ML) and Deep Learning (DL)
MRI has made immense strides in utilizing new and innovative imaging techniques to maximize the information measured from a patient with this non-invasive and low-harm modality. New endeavors currently being explored, and the future of MRI, include but are not limited to the use of artificial intelligence (AI), machine learning (ML), and deep learning (DL) in imaging, as well as the prospects of magnetic resonance spectroscopy (MRS). Artificial intelligence refers to machines that respond to stimulation with the human capacity of contemplation, judgment, and intention. ML is AI that learns from the environment and using a variety of algorithms, improves data, spots patterns, and performs actions on those patterns. DL is ML that uses neural networks instead of human intervention to obtain data and make predictions/actions and can function/observe data at many different levels. We speculate that combining MRI with these techniques could possibly lead to microscopic resolution [32].
Currently, AI, ML, and DL are being used in a variety of innovative ways to maximize the benefits technology gives us in MR imaging. Some of these applications involve ML being used to assess radiomics (qualitative image data being converted into quantitative data) of disease progression and early cancer diagnosis. For example, ML has been able to assist radiologists in determining the malignancy of lesions in patients with prostate cancer [33]. Additionally, it has been used to project the effectiveness of neoadjuvant chemotherapy (NAC) as a treatment for patients with breast cancer, and then determine increases/decreases in treatment load based on the predicted prognosis and patient reaction to the treatment [34]. DL has been used to convert low-resolution images obtained with low-field strength MRI machines to high-resolution high-field strength images, giving a much higher level of detail and information without the cost of the high-field strength machinery. Furthermore, ML is being used to synthesize full-contrast medium MR images from no or low-contrast patient images [35]. Additionally, AI and specifically DL have been used to correct for large, complicated motion artifacts and field inhomogeneities [36]. These are only some of the applications of this technology to the field of MR imaging, an initiative that continues to grow and holds immense potential and hope for the future.
Magnetic resonance spectroscopy
- Recording the spectra from multiple regions of the brain and using that to map out the spatial distribution of metabolites in the brain is beneficial when dealing with tumors' metabolic inhomogeneities mainly because the spectra from the necrotic core of high-grade tumor is very different from that of an actively growing rim.
- Differentiate between tumor type and also tumor grade in conjunction with ASL by analyzing choline levels in areas of high cerebral blood flow.
- Recognize high metabolic activity and construct Cho maps in order to determine active tumor and tumor growth regions which help in targeted radiation therapy [40].
Other
Some other technologies currently being researched to advance MRI include (but are not limited to):
- X-nuclei imaging: Focuses on the detection of nuclei of other atoms such as sodium, potassium, etc. in tissues. Although this is still in clinical trials, it aims to reveal the underlying changes in the physiological processes on a cellular level. This also shows us the feasibility of combining UHF MRI with other imaging modalities for improved image quality [41].
- Parallel RF Transmission: It decreases the inhomogeneities of RF excitations and RF energy depositions which usually limit some spectral resolution. Its main goal is to smooth the RF pulse to the region of interest so as to get a better signal. It uses different RF pulse sequences at the same time [42].
- Motion correction: Improves image quality and reduces movement artifacts [31].
- Amide Proton Transfer Weighted Imaging (APTw): APTw is a subset of Chemical Exchange Saturation Transfer (CEST) MRI where set radiofrequency pulses preferentially excite the population of protons in the amide groups of target proteins (such as surface proteins of cancer cells). Eventually, there will be a saturated population of excited protons. These exciting protons are continuously exchanged with protons from circulating water molecules. So many saturated protons are exchanged that there will be a noticeable change in the MR signal from the bulk population of water molecules. Given that this approach is sensitive and quantifiable, the extent of this drop in bulk signal reflects the size and density of the target tissue [43,44]. This technique has been known to distinguish between low and high-grade gliomas [45].
Diagnostic/Clinical MRI
Throughout this review, we’ve established that UHF MRI enables the collection of higher-resolution images due to an increased signal-to-noise ratio. While most clinical MRI machines are currently 3T, the prospects of 7T and 11.7T MRI machines are being discussed from the clinical perspective. As an example, the Richard M. Lucas Center for Imaging at Stanford University has inaugurated a 7T clinical MRI dedicated to whole-body scanning [46]. Such higher-quality images have shown great benefits in neuroimaging in particular, specifically in conditions such as multiple sclerosis (MS), cerebrovascular disease, epilepsy, gliomas/neuro-oncology, and neurodegenerative diseases [47]. Higher resolution images enable physicians to qualify white and gray matter lesions with increased accuracy and improve the detection of metabolic markers/metabolites (in the case of MS and cerebrovascular disease). Furthermore, UHF MRI would enable better visualization of the changes in cerebral cortical structure in neurodegenerative disease and neuro-oncology, even helping visualize the smaller and smaller vessels for stroke detection and cerebral vascularization mapping [47,48].
Improved spatial resolution will not only significantly advance the study of cerebral anatomy, but will also enhance the pre-surgical determination of tumor extraction sites in cases of neuro-oncology. Magnetic resonance spectroscopy and other techniques can be used to assess tumor grade and treatment efficacy with greater precision. In cases of glioma and other neuro-oncologic pathologies, higher resolution can also be used to detect microbleeds, visualize intratumoral structures and neovascularization, and specifically localize the targeted areas for radiotherapy [49].
While only the applications of UHF MRI in neuroimaging have been explored here, with the immense variety of imaging techniques and growing research in UHF MRI, hopefully, its immense benefit to the clinical toolkit will be increasingly acknowledged, and the barriers to its implementation will be surpassed by the incorporation of both Artificial Intelligence and machine learning technologies.
Conclusion
Magnetic Resonance Imaging, otherwise known as MRI, is an imaging technique that all physicians, researchers, medical personnel, and a large majority of the general population have at least heard of. However, how many truly know the science and reasoning behind its functionality? The purpose of this review is to first thoroughly understand and explore the functionality of MRI, and the methodology behind it. Then, the question of whether microscopic imaging is possible today with current ultrahigh field strength scanners was explored, yielding the conclusion that as of now, mesoscopic imaging is possible with various imaging techniques, but the microscopic resolution will still require advancement in imaging hardware, in more refined scanning sequences and the incorporation of novel artificial intelligence protocols.
A detailed comparison of 1.5 T, 3 T, and 7T MRI scanners show a clear correlation between increasing magnetic field strength and improved image resolution. However, such advances in hardware must be complemented by various novel imaging sequences that can produce higher-resolution images or images that give us particular information about the tissue. Diffusion-weighted imaging (DWI) characterizes the movement of water molecules in tissues to map out details of structures and identify areas of abnormal tissue. Pulse gradient spin echo (PGSE) and oscillating gradient spin echo (OGSE) are pulse sequences used within DWI imaging to gain greater detail regarding diffusive properties of tissues, utilizing two separate and superimposed images (PGSE) or an added dimension of data collection (OGSE). Functional MRI (fMRI) via arterial spin labeling (ASL) and blood oxygen level-dependent (BOLD) techniques employ an endogenous contrast to better visualize microcirculation and vessel structure. ASL involved magnetically differentiating/tagging moving protons of water molecules in the blood. BOLD works by detecting the slight magnetic changes in oxygenated and deoxygenated hemoglobin in the blood, identifying areas of higher blood flow and activity. The techniques summarized here are merely a few of the sequences and imaging techniques being used/studied in radiology.
Such approaches in UHF-MRI are particularly useful in neurology [50], where diffusion MRI studies have led to numerous functional insights into the brain [51-53]. Functional studies of the brain have been complemented by structural studies of the brain because of explorations into pushing the resolution of UHF-MRI towards the mesoscopic scale of resolution [54]. These studies in turn complement computer tomographic (CT) research into brain anatomy [55]. Further insights should be gained by newer more sensitive protocols such as APTw MRI, an approach known to be optimized in higher field strength MRI [43-45]. Also, diffusion-weighted protocols such as OGSE have the potential to sense variations in the intracellular structure; a feature that should be enhanced with higher field strength MRI [56]. OGSE signals are also sensitive to axonal diameter [57] thus lending the approach useful in assessing neuronal network anatomy.
While these techniques have launched us forward in clinical imaging capabilities today, there are still many that are being studied today. Some of the primary techniques and technologies being studied include, but aren’t limited to, the use of machine learning (ML), deep learning (DL), and artificial intelligence (AI) in conjugation with MRI [55,58-61]. These technologies are being used to assess radiomic data from images, as predictive software to project disease prognoses, to convert low-resolution images to high-resolution ones, and to correct motion artifacts while scanning. Magnetic resonance spectroscopy (MRS) is being used to create metabolite maps, to determine tumor size, location, and grade, as well as to recognize/classify the metabolic activity of tumors. In addition, X-nuclei imaging (detection of ion nuclei to study physiologic changes), parallel RF transmission (controlling for RF field inhomogeneities), and motion correction are all initiatives that are being studied further.
MRI has immense potential to serve as the primary and most effective soft tissue imaging modality. Its applications and capabilities seem to increase with every study, and there is still much to learn in terms of how to optimize and advance this imaging process. Hopefully, with more studies, MRI will be able to minimize the amount of ionizing radiation used in medicine and help propel imaging and diagnostics for cancer and other ailments into a (literally) brighter future.
We thank James Strommer for his excellent scientific illustrations in Figures 1,2. We thank Drs. Heike Daldrup-Link, Chirag Patel, and Wei Wu for their invaluable intellectual and editorial insights.
- Schild HH. MRI Made Easy (well almost). Schering AG Berlin 1990.
- Hopkins J. Bloomberg School of Public Health (August, 24, 2015). Principles of fMRI Part 1, Module 7: K-space. YouTube. https://www.youtube.com/watch?v=FI5frNsRTI4&list=PLfXA4opIOVrGHncHRxI3Qa5GeCSudwmxM&index=8.
- Jhamb TK, Rejathalal V, Govindan VK. A Review on Image Reconstruction through MRI k-Space Data. Mod Educ Comput Sci. 2015; 7:42–59.
- Moratal D, Vallés-Luch A, Martí-Bonmatí L, Brummer M. k-Space tutorial: an MRI educational tool for a better understanding of k-space. Biomed Imaging Interv J. 2008 Jan;4(1):e15. doi: 10.2349/biij.4.1.e15. Epub 2008 Jan 1. PMID: 21614308; PMCID: PMC3097694.
- LOFT Lab. Introduction to k-Space. YouTube. 2020. https://www.youtube.com/watch?v=GF7Z8Sd9qYE
- Icahn School of Medicine. K-Space: A way to understand how MRI parameters affect images. YouTube. 2016. https://www.youtube.com/watch?v=QHtZR0mtB80
- Weldon KB, Olman CA. Forging a path to mesoscopic imaging success with ultra-high field functional magnetic resonance imaging. Philos Trans R Soc Lond B Biol Sci. 2021 Jan 4;376(1815):20200040. doi: 10.1098/rstb.2020.0040. Epub 2020 Nov 16. PMID: 33190599; PMCID: PMC7741029.
- Magee T, Shapiro M, Williams D. Comparison of high-field-strength versus low-field-strength MRI of the shoulder. AJR Am J Roentgenol. 2003 Nov;181(5):1211-5. doi: 10.2214/ajr.181.5.1811211. PMID: 14573405.
- Tocchio S, Kline-Fath B, Kanal E, Schmithorst VJ, Panigrahy A. MRI evaluation and safety in the developing brain. Semin Perinatol. 2015 Mar;39(2):73-104. doi: 10.1053/j.semperi.2015.01.002. Epub 2015 Mar 3. PMID: 25743582; PMCID: PMC4380813.
- Boer VO, Klomp DW, Juchem C, Luijten PR, de Graaf RA. Multislice ¹H MRSI of the human brain at 7 T using dynamic B₀ and B₁ shimming. Magn Reson Med. 2012 Sep;68(3):662-70. doi: 10.1002/mrm.23288. Epub 2011 Dec 12. PMID: 22162089; PMCID: PMC3306521.
- Elster AD. Advantages of Lower Field Scanners. Questions and Answers in MRI. (2015). http://mriquestions.com/advantages-to-low-field.html.
- Elster AD. Low field Disadvantages. Questions and Answers in MRI. (2015). http://mriquestions.com/disadvantages.html
- Erturk MA, Li X, Van de Moortele PF, Ugurbil K, Metzger GJ. Evolution of UHF Body Imaging in the Human Torso at 7T: Technology, Applications, and Future Directions. Top Magn Reson Imaging. 2019 Jun;28(3):101-124. doi: 10.1097/RMR.0000000000000202. PMID: 31188271; PMCID: PMC6587233.
- Fujiwara S, Mori Y, de la Mora DM, Akamatsu Y, Yoshida K, Shibata Y, Masuda T, Ogasawara K, Yoshioka Y. Feasibility of IVIM parameters from diffusion-weighted imaging at 11.7T MRI for detecting ischemic changes in common carotid artery occlusion rats. Sci Rep. 2020 May 21;10(1):8404. doi: 10.1038/s41598-020-65310-8. PMID: 32439877; PMCID: PMC7242437.
- Adil SM, Calabrese E, Charalambous LT, Cook JJ, Rahimpour S, Atik AF, Cofer GP, Parente BA, Johnson GA, Lad SP, White LE. A high-resolution interactive atlas of the human brainstem using magnetic resonance imaging. Neuroimage. 2021 Aug 15;237:118135. doi: 10.1016/j.neuroimage.2021.118135. Epub 2021 May 2. PMID: 33951517; PMCID: PMC8480283.
- CEA Recherche (November, 19, 2021). World premiere the most powerful MRI scanner in the world delivers its first images!. YouTube. https://www.youtube.com/watch?v=N6CJy2uyKlQ
- Callewaert B, Jones EAV, Himmelreich U, Gsell W. Non-Invasive Evaluation of Cerebral Microvasculature Using Pre-Clinical MRI: Principles, Advantages and Limitations. Diagnostics (Basel). 2021 May 21;11(6):926. doi: 10.3390/diagnostics11060926. PMID: 34064194; PMCID: PMC8224283.
- Elster AD. Spin Echo. Questions and Answers in MRI. (2015). http://mriquestions.com/spin-echo1.html .
- Chilla GS, Tan CH, Xu C, Poh CL. Diffusion weighted magnetic resonance imaging and its recent trend-a survey. Quant Imaging Med Surg. 2015 Jun;5(3):407-22. doi: 10.3978/j.issn.2223-4292.2015.03.01. PMID: 26029644; PMCID: PMC4426106.
- Fink KRT, Fink JR. Principles of Modern Neuroimaging. Princ. Neurol. Surg., n.d., 62–86.
- Szafer A, Zhong J, Anderson AW, Gore JC. Diffusion-weighted imaging in tissues: theoretical models. NMR Biomed. 1995 Nov-Dec;8(7-8):289-96. doi: 10.1002/nbm.1940080704. PMID: 8739267.
- DWI Pulse Sequence n.d.
- Xu J, Jiang X, Li H, Arlinghaus LR, McKinley ET, Devan SP, Hardy BM, Xie J, Kang H, Chakravarthy AB, Gore JC. Magnetic resonance imaging of mean cell size in human breast tumors. Magn Reson Med. 2020 Jun;83(6):2002-2014. doi: 10.1002/mrm.28056. Epub 2019 Nov 25. PMID: 31765494; PMCID: PMC7047520.
- Kershaw J, Obata T. Oscillating-gradient spin-echo diffusion-weighted imaging (OGSE-DWI) with a limited number of oscillations: I. Signal equation. J Magn Reson. 2021 May;326:106962. doi: 10.1016/j.jmr.2021.106962. Epub 2021 Mar 9. PMID: 33756275.
- Padron Olivas MF. Oscillating Gradient Spin Echo (OGSE): A Study of Short Diffusion Time Effects in Human Brain at 3T. Univ Alta 2020.
- Petcharunpaisan S, Ramalho J, Castillo M. Arterial spin labeling in neuroimaging. World J Radiol. 2010 Oct 28;2(10):384-98. doi: 10.4329/wjr.v2.i10.384. PMID: 21161024; PMCID: PMC2999014.
- Dury RJ, Falah Y, Gowland PA, Evangelou N, Bright MG, Francis ST. Ultra-high-field arterial spin labelling MRI for non-contrast assessment of cortical lesion perfusion in multiple sclerosis. Eur Radiol. 2019 Apr;29(4):2027-2033. doi: 10.1007/s00330-018-5707-5. Epub 2018 Oct 2. PMID: 30280247; PMCID: PMC6420612.
- Forster BB, MacKay AL, Whittall KP, Kiehl KA, Smith AM, Hare RD, Liddle PF. Functional magnetic resonance imaging: the basics of blood-oxygen-level dependent (BOLD) imaging. Can Assoc Radiol J. 1998 Oct;49(5):320-9. PMID: 9803232.
- Neugarten J, Golestaneh L. Blood oxygenation level-dependent MRI for assessment of renal oxygenation. Int J Nephrol Renovasc Dis. 2014 Nov 21;7:421-35. doi: 10.2147/IJNRD.S42924. PMID: 25473304; PMCID: PMC4247132.
- Vincent K, Moore J, Kennedy S, Tracey I. Blood oxygenation level dependent functional magnetic resonance imaging: current and potential uses in obstetrics and gynaecology. BJOG. 2009 Jan;116(2):240-6. doi: 10.1111/j.1471-0528.2008.01993.x. PMID: 19076956; PMCID: PMC2675013.
- Ladd ME, Bachert P, Meyerspeer M, Moser E, Nagel AM, Norris DG, Schmitter S, Speck O, Straub S, Zaiss M. Pros and cons of ultra-high-field MRI/MRS for human application. Prog Nucl Magn Reson Spectrosc. 2018 Dec;109:1-50. doi: 10.1016/j.pnmrs.2018.06.001. Epub 2018 Jun 8. PMID: 30527132.
- Kavlakoglu E. AI vs. Machine Learning vs. Deep Learning vs. Neural Networks: What’s the Difference? 2020.
- Cuocolo R, Cipullo MB, Stanzione A, Ugga L, Romeo V, Radice L, Brunetti A, Imbriaco M. Machine learning applications in prostate cancer magnetic resonance imaging. Eur Radiol Exp. 2019 Aug 7;3(1):35. doi: 10.1186/s41747-019-0109-2. PMID: 31392526; PMCID: PMC6686027.
- Lo Gullo R, Eskreis-Winkler S, Morris EA, Pinker K. Machine learning with multiparametric magnetic resonance imaging of the breast for early prediction of response to neoadjuvant chemotherapy. Breast. 2020 Feb;49:115-122. doi: 10.1016/j.breast.2019.11.009. Epub 2019 Nov 23. PMID: 31786416; PMCID: PMC7375548.
- Hu H. Recent Advances of Bioresponsive Nano-Sized Contrast Agents for Ultra-High-Field Magnetic Resonance Imaging. Front Chem. 2020 Mar 20;8:203. doi: 10.3389/fchem.2020.00203. PMID: 32266217; PMCID: PMC7100386.
- Tamada D. Review: Noise and artifact reduction for MRI using deep learning. Magn Reson Med Sci 2020.
- Mayfield Brain and Spine. Magnetic Resonance (MR) Spectroscopy. 2018. https://mayfieldclinic.com/pe-mrspectroscopy.htm
- Saber M, Gaillard F. (May, 4, 2022). MR Spectroscopy. Radiopaedia. https://radiopaedia.org/articles/mr-spectroscopy-1?lang=us#:~:text=References-,Physics,return%20a%20slightly%20different%20signal.
- Godlewska BR, Clare S, Cowen PJ, Emir UE. Ultra-High-Field Magnetic Resonance Spectroscopy in Psychiatry. Front Psychiatry. 2017 Jul 11;8:123. doi: 10.3389/fpsyt.2017.00123. PMID: 28744229; PMCID: PMC5504194.
- Horská A, Barker PB. Imaging of brain tumors: MR spectroscopy and metabolic imaging. Neuroimaging Clin N Am. 2010 Aug;20(3):293-310. doi: 10.1016/j.nic.2010.04.003. PMID: 20708548; PMCID: PMC2927327.
- Hu R, Kleimaier D, Malzacher M, Hoesl MAU, Paschke NK, Schad LR. X-nuclei imaging: Current state, technical challenges, and future directions. J Magn Reson Imaging. 2020 Feb;51(2):355-376. doi: 10.1002/jmri.26780. Epub 2019 May 17. PMID: 31102340.
- Katscher U, Börnert P. Parallel RF transmission in MRI. NMR Biomed. 2006 May;19(3):393-400. doi: 10.1002/nbm.1049. PMID: 16705630.
- Kamimura K, Nakajo M, Yoneyama T, Takumi K, Kumagae Y, Fukukura Y, Yoshiura T. Amide proton transfer imaging of tumors: theory, clinical applications, pitfalls, and future directions. Jpn J Radiol. 2019 Feb;37(2):109-116. doi: 10.1007/s11604-018-0787-3. Epub 2018 Oct 19. PMID: 30341472.
- Wu B, Warnock G, Zaiss M, Lin C, Chen M, Zhou Z, Mu L, Nanz D, Tuura R, Delso G. An overview of CEST MRI for non-MR physicists. EJNMMI Phys. 2016 Dec;3(1):19. doi: 10.1186/s40658-016-0155-2. Epub 2016 Aug 26. PMID: 27562024; PMCID: PMC4999387.
- Suh CH, Park JE, Jung SC, Choi CG, Kim SJ, Kim HS. Amide proton transfer-weighted MRI in distinguishing high- and low-grade gliomas: a systematic review and meta-analysis. Neuroradiology. 2019 May;61(5):525-534. doi: 10.1007/s00234-018-02152-2. Epub 2019 Jan 21. PMID: 30666352.
- The Richard M. Lucas Center for Imaging 2022.
- Barisano G, Sepehrband F, Ma S, Jann K, Cabeen R, Wang DJ, Toga AW, Law M. Clinical 7 T MRI: Are we there yet? A review about magnetic resonance imaging at ultra-high field. Br J Radiol. 2019 Feb;92(1094):20180492. doi: 10.1259/bjr.20180492. Epub 2018 Nov 1. PMID: 30359093; PMCID: PMC6404849.
- Trattnig S, Springer E, Bogner W, Hangel G, Strasser B, Dymerska B, Cardoso PL, Robinson SD. Key clinical benefits of neuroimaging at 7T. Neuroimage. 2018 Mar;168:477-489. doi: 10.1016/j.neuroimage.2016.11.031. Epub 2016 Nov 13. PMID: 27851995; PMCID: PMC5832016.
- Shaffer A, Kwok SS, Naik A, Anderson AT, Lam F, Wszalek T, Arnold PM, Hassaneen W. Ultra-High-Field MRI in the Diagnosis and Management of Gliomas: A Systematic Review. Front Neurol. 2022 Apr 5;13:857825. doi: 10.3389/fneur.2022.857825. PMID: 35449515; PMCID: PMC9016277.
- Taghizadeh S, Labuda C, Yang CC, Morris B, Kanakamedala MR, Vijayakumar S, Rey-Dios R, Duggar WN, Florez E, Fatemi A. Optimizing MRI sequences and images for MRI-based stereotactic radiosurgery treatment planning. Rep Pract Oncol Radiother. 2019 Jan-Feb;24(1):12-19. doi: 10.1016/j.rpor.2018.09.010. Epub 2018 Oct 10. PMID: 30337843; PMCID: PMC6187087.
- Deelchand DK, Ho ML, Nestrasil I. Ultra-High-Field Imaging of the Pediatric Brain and Spinal Cord. Magn Reson Imaging Clin N Am. 2021 Nov;29(4):643-653. doi: 10.1016/j.mric.2021.06.013. PMID: 34717851.
- Gallichan D. Diffusion MRI of the human brain at ultra-high field (UHF): A review. Neuroimage. 2018 Mar;168:172-180. doi: 10.1016/j.neuroimage.2017.04.037. Epub 2017 Apr 18. PMID: 28428047.
- Uludağ K, Blinder P. Linking brain vascular physiology to hemodynamic response in ultra-high field MRI. Neuroimage. 2018 Mar;168:279-295. doi: 10.1016/j.neuroimage.2017.02.063. Epub 2017 Feb 22. PMID: 28254456.
- Dumoulin SO, Fracasso A, van der Zwaag W, Siero JCW, Petridou N. Ultra-high field MRI: Advancing systems neuroscience towards mesoscopic human brain function. Neuroimage. 2018 Mar;168:345-357. doi: 10.1016/j.neuroimage.2017.01.028. Epub 2017 Jan 16. PMID: 28093360.
- Xue G, Chen C, Lu ZL, Dong Q. Brain Imaging Techniques and Their Applications in Decision-Making Research. Xin Li Xue Bao. 2010 Feb 3;42(1):120-137. doi: 10.3724/SP.J.1041.2010.00120. PMID: 20376329; PMCID: PMC2849100.
- Xu J, Does MD, Gore JC. Sensitivity of MR diffusion measurements to variations in intracellular structure: effects of nuclear size. Magn Reson Med. 2009 Apr;61(4):828-33. doi: 10.1002/mrm.21793. PMID: 19205020; PMCID: PMC2749035.
- Drobnjak I, Zhang H, Ianuş A, Kaden E, Alexander DC. PGSE, OGSE, and sensitivity to axon diameter in diffusion MRI: Insight from a simulation study. Magn Reson Med. 2016 Feb;75(2):688-700. doi: 10.1002/mrm.25631. Epub 2015 Mar 25. PMID: 25809657; PMCID: PMC4975609.
- Zhang Z, Li G, Xu Y, Tang X. Application of Artificial Intelligence in the MRI Classification Task of Human Brain Neurological and Psychiatric Diseases: A Scoping Review. Diagnostics (Basel). 2021 Aug 3;11(8):1402. doi: 10.3390/diagnostics11081402. PMID: 34441336; PMCID: PMC8392727.
- Thaha MM, Kumar KPM, Murugan BS, Dhanasekeran S, Vijayakarthick P, Selvi AS. Brain Tumor Segmentation Using Convolutional Neural Networks in MRI Images. J Med Syst. 2019 Jul 24;43(9):294. doi: 10.1007/s10916-019-1416-0. PMID: 31342192.
- Hoseini F, Shahbahrami A, Bayat P. An Efficient Implementation of Deep Convolutional Neural Networks for MRI Segmentation. J Digit Imaging. 2018 Oct;31(5):738-747. doi: 10.1007/s10278-018-0062-2. PMID: 29488179; PMCID: PMC6148810.
- He X, Xu W, Yang J, Mao J, Chen S, Wang Z. Deep Convolutional Neural Network With a Multi-Scale Attention Feature Fusion Module for Segmentation of Multimodal Brain Tumor. Front Neurosci. 2021 Nov 26;15:782968. doi: 10.3389/fnins.2021.782968. PMID: 34899175; PMCID: PMC8662724.
- Baliyan V, Das CJ, Sharma R, Gupta AK. Diffusion weighted imaging: Technique and applications. World J Radiol. 2016 Sep 28;8(9):785-798. doi: 10.4329/wjr.v8.i9.785. PMID: 27721941; PMCID: PMC5039674.
- Drobnjak I, Zhang H, Ianus A, Kaden E, Alexander D, OGSE P. and sensitivity to axon diameter in diffusion MRI: Insight from a simulation study. Magn Reson Med 2015;75:688–700.
- Ferré JC, Bannier E, Raoult H, Mineur G, Carsin-Nicol B, Gauvrit JY. Arterial spin labeling (ASL) perfusion: techniques and clinical use. Diagn Interv Imaging. 2013 Dec;94(12):1211-23. doi: 10.1016/j.diii.2013.06.010. Epub 2013 Jul 11. PMID: 23850321.
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.





 Save to Mendeley
Save to Mendeley
