Journal of Dental Problems and Solutions
Autophagy and the potential linkage with the human oral diseases
Deniz Yaman1,2, Hasibe Verdi1,3 and F Belgin Ataç1,3*,
2Department of Oral and Maxillofacial Surgery, Bolu Abant Izzet Baysal University Faculty of Dentistry, Bolu, Turkey
3Department of Medical Biology, Başkent University Faculty of Medicine, Ankara, Turkey
Cite this as
Yaman D, Verdi H, Ataç FB (2020) Autophagy and the potential linkage with the human oral diseases. J Dent Probl Solut 7(1): 010-019. DOI: 10.17352/2394-8418.000078Autophagy is a cell survival mechanism that has a key role in both physiologic and pathologic conditions. In this cellular ‘clearance’ process, damaged organelles and macromolecules are transported to the lysosomes for degradation. With autophagy, a protection mechanism occurs in the cell under difficult conditions including oxidative stress and lack of growth factor. Paradoxically, in autophagy under some conditions, non-apoptotic cell death can be observed via a caspase-independent pathway. Recent, studies indicated that there is a strong correlation between the composition of the oral microbiota and the pathogenesis of oral diseases, and autophagy plays a crucial role in the development of this process. This review is focused on the molecular mechanism of autophagy and its relationship with human oral diseases. In this review, the importance of the autophagy mechanism in the maintenance of oral tissue health is emphasized. After examining the studies, we observed the importance of autophagy in response to a variety of oral diseases and aimed to present this awareness. Thus, a better understanding of autophagy will be the basis for developing new therapeutic strategies in the treatment of oral diseases.
Autophagy
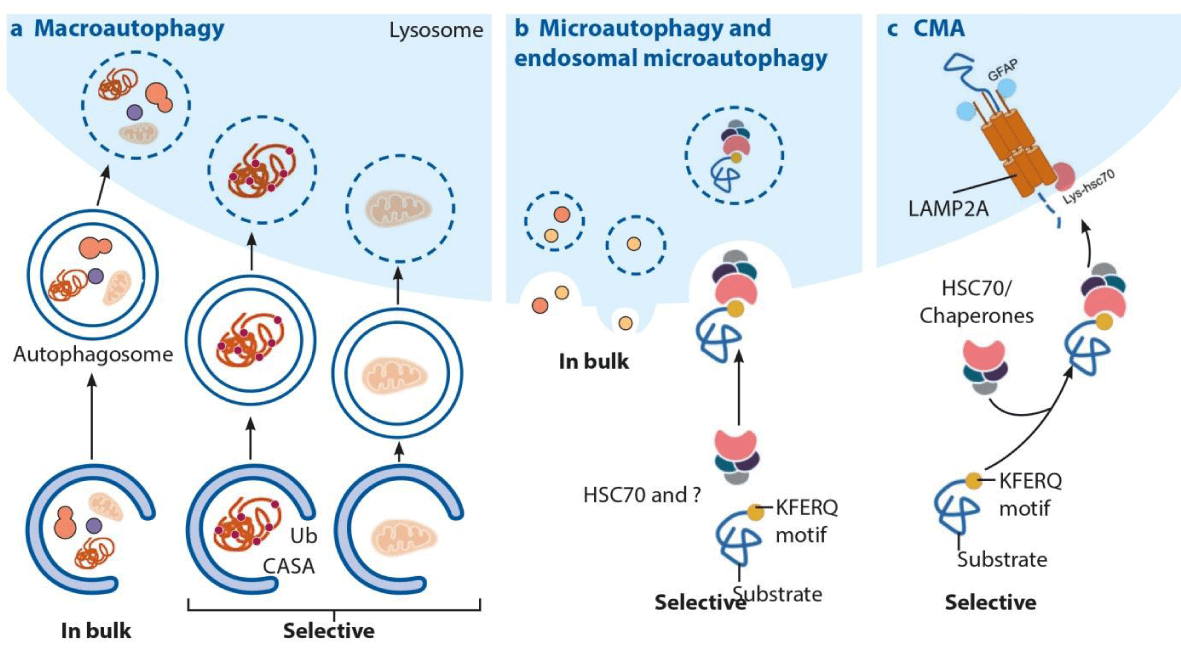
Autophagy is the basic physiological process responsible for the degradation of damaged organelles, toxic protein aggregates, intracellular bacterial or viral pathogens [1-3]. In all eukaryotic cells with autophagy, nutrients are recycled by providing alternative energy for cell metabolism under conditions such as starvation, heat, inflammation, hypoxia and oxidative stress [4]. Today, three different types of autophagy pathways have been defined as micro-autophagy, macro-autophagy and chaperone-mediated autophagy [5,6] (Figure 1).
Proteins to be degraded by Chaperone-Mediated Autophagy (CMA) contain a unique motif that is biochemically associated with the KFERQ (Figure 1) [6,7]. When the protein is not correctly folded or damaged, this motif is revealed. It is recognized by a molecular chaperone called Hsc70 (Heat shock cognate protein 70). Hsc70 binds to this unique motif and directs the protein to the lysosomal surface by forming the substrate-Hsc70 complex. The lysosomal surface has a protein called Lysosomal Membrane Protein 2A (LAMP-2A). This protein acts as a receptor for the substrate-Hsc70 complex [8]. LAMP-2A creates structural changes to form a hollow, cylindrical transport structure called the CMA translocation complex. The unfolded substrate passes through this translocation complex and enters the lysosomal lumen. After the substrate enters the lysosomal lumen, the CMA translocation complex is immediately disintegrated by Hsc70 and other proteins in the lysosomal membrane. The substrate is degraded by proteases in the lumen and amino acids are released into the cytosol [6,9] (Figure 1). In micro-autophagy; the cytosolic components pass directly into the lysosome by fusing with the lysosome membrane [6,9,10] (Figure 1).
Macro-autophagy; hereafter referred to as autophagy, is one of the most studied and therefore the most known molecular details of autophagy. In this autophagy; formation of the phagophore (isolation membrane surrounding the cytoplasmic components), autophagosome (double-membrane vacuole formed by phagophore elongation) and autolysosome (autophagosome and lysosome fusion for degradation of cytosolic components) are important [11,12] (Figure 2).
Many related genes and regulatory proteins are involved in the activation of autophagy. In nutrient deficiency; AMP-Activated Protein Kinase (AMPK), an energy hemostasis sensor, triggers autophagosome formation. AMPK; inhibits the mammalian target of rapamycin complex 1 (mTORC1), which is the general suppressor of autophagy. Besides, AMPK activates the autophagosome-initiating complex such as Unc-51-like kinase 1 (ULK1), class III phosphatidylinositol 3 kinase (PI3 K) [13,14]. The class III PI3K/PtdIns(3) K complex is consists of ‘vacuolar protein sorting 34 (VPS34/ PIK3C3), Vps15 (PIK3R4), Beclin-1 and Atg14. This complex is important for the phosphatidylinositol 3-phosphate (PI(3)P) production and autophagosomal membrane formation [15]. The increase in PI(3)P concentration is a precursor in the formation of autophagosomes and it triggers the accumulation of proteins associated with autophagy. Also, intracellular two different molecular systems take part in the elongation of autophagosome membrane. They are known as ‘Atg5-Atg12- Atg16’ and ‘microtubule-associated protein light chain 3 isoform I (LC3I)’ systems [16,17]. With the Atg5-Atg12- Atg16 complex, the vesicle is enlarged [18]. The microtubule-associated protein light chain 3 isoform II (LC3II) acts as a link for many proteins involved in autophagy and is responsible for elongation and closure of the phagophore membrane. Besides, the increase in the ratio of LC3II / LC3I shows an increase in the number of autophagosomes, so it is an important marker that should be examined in the studies [19-21]. Degradation of cytosolic components, which is the last step of autophagy, is completed by the fusion of the autophagosome with lysosomal membrane (autolysosome) [22]. After lysosome and autophagosome junction; cytosolic components are broken down by hydrolytic enzymes and they are released back into the cytoplasm for use in the re-synthesis of biological molecules (Figure 2)[12].
Recent studies have shown that intracellular autophagy disruptions contribute to disease formation and progression [23]. Various chemical agents have provided us with detailed information about the effect of autophagy on the progression of some diseases. From these chemical agents, 3-methylamine (3-MA) and Bafilomycin A1 are known to inhibit autophagy, but rapamycin and lithium are known to induce autophagy (Figure 2)[12]. With the help of these chemical agents, previous studies have demonstrated that autophagy is an endogenous defense mechanism for cells to regulate multiple cellular pathways involved in anti-aging, tumor suppression and promotion, inflammation, and immune response [24-26]. In this review, the effect of autophagy on bone resorption, inflammation of periodontal tissues and oral cancer progression was evaluated.
Autophagy and bone resorption
Molecular mechanisms involved in the formation of the bone structure of the craniofacial region; differ in terms of the organization and embryonic origin from other parts of the skeleton [27]. Any deterioration in this unique structure can lead to craniofacial dysmorphology and associated morbidities [28]. In previous studies, various factors affecting the development of craniofacial skeletal structure was reported [29,30]. Besides, the role of autophagy on craniofacial skeletal development and regulatory mechanisms has been examined in the studies. Thomas et al. shared the results of their study investigating the effects of Fip200 and Atg5 deletion on craniofacial skeletal development [27]. According to this; it was determined that osteoblast mineralization was negatively affected and osteopenia was observed due to autophagy suppression [27]. Onal, et al. demonstrated the suppression of autophagy via deletion of Atg7 gene in osteocytes of 6-month-old mice [31]. Also, they found that cancellous bone volume and cortical thickness decreased and cortical porosity increased. These results showed that both Atg5 and Atg7 genes are important for phagophore formation in autophagy and normal bone metabolism.
Bone is a dynamic tissue that undergoes continuous remodeling due to osteoblast-mediated bone formation and osteoclast-mediated bone resorption [32]. Producing and mineralizing the bone matrix is one of the basic functions of osteoblasts [33]. Inadequate autophagic activation in osteoblasts due to the accumulation of oxidative stress associated with advanced age is a recognized fact [33]. According to the study of Nollet et al. the induction of autophagy was associated with increased bone matrix mineralization[33]. After inhibition of autophagy in osteoblast cells, a decrease in bone mineralization capacity was reported [33].
As known, osteoporosis is a condition characterized by low Bone Mineral Density (BMD) and an increased risk of fracture [34]. A decrease in the production of growth factors or estrogen hormone (especially in postmenopausal women) are one of the extrinsic mechanism and they are associated with osteoporosis in advanced age [35]. Besides, long-term corticoid treatment, inadequate physical exercise also contributes to the development of osteoporosis [36,37]. Currently, autophagy has been suggested to be an intrinsic mechanism affecting the survival of osteoblasts, osteoclasts, and chondrocytes in hypoxia [38]. Li et al. investigated the effect of Atg7 during the developmental and remodeling stages of bone in vivo [35]. They found that Atg 7 deficiency triggered endoplasmic reticulum (ER) stress in osteoblasts [35]. Also, decreased bone mass and mineralization in both stages were observed [35]. Altogether, the studies demonstrated the protective role of autophagy on bone metabolism.
Autophagy and periodontal diseases
The periodontium is the specialized tissue that both surrounds and supports the teeth, dissipates the forces generated during chewing-speaking-swallowing and preserve the integrity of oral mucosa. The gingiva, periodontal ligament, cementum and alveolar bone components that make up this structure exhibit morphological and functional integrity [39]. Periodontal disease develops with chronic bacterial inflammation of these tissues that make up the periodontium. Destruction of the periodontium due to chronic inflammation is the most important cause of tooth loss in adults [40]. Although the primary cause of periodontal diseases is bacterial plaque, there is a consensus that some systemic and metabolic factors increase the risk of periodontitis by affecting disease severity and prognosis [39,41]. Smoking, poor oral hygiene, hyperglycemia, occlusal trauma, osteoporosis, Chediak-Higashi syndrome, cyclic neutropenias, agranulocytosis, leukocyte adhesion deficiency and Papillon Lefevre syndrome are potential risk factors for the development of periodontitis [42].
Gram-negative bacteria such as Bacteroides forsythus, Aggregatibacter actinomycetemcomitans (A.a) and Porphyromonas gingivalis (Pg) are well-established periodontal pathogens, and existence great numbers of these bacteria in the sulcular epithelium is the first step for the development of periodontal diseases [3, 43]. Any deficiency in the attached keratinized gingiva would hamper to maintain efficient oral hygiene and lead the penetration of microbial plaque into the gingival crevice which would trigger the activation of neutrophils, monocyte/macrophage, B and T cells around the inflamed tissue [44,45]. Overresponsive monocyte/macrophage, B and T cells phenotyping in patients with periodontitis causes excessive production of catabolic factors and increases the destruction of connective tissue and bone [46]. Bullon et al. investigated the role of autophagy in peripheral blood mononuclear cells from patients with periodontitis [47]. They found that patients with periodontitis have a higher level of Autophagy-Related Genes (ATGs) in peripheral blood mononuclear cells compared to healthy participants [47]. Chung, et al. investigated the anti-inflammatory effect of Trans-Cinnamic Aldehyde (TCA) against Aggregatibacter actinomycetemcomitans (Aa) infection in human macrophages and mice [48]. After TCA treatment, it was found that the release of proinflammatory mediators such as tumor necrosis factor-𝛼 and interleukin (IL)-1beta decreased [48]. These proinflammatory mediators are involved in the destruction of periodontal tissues, and their decreased levels have shown that bone loss is inhibited [48]. Also, the autophagosome formation and the expressions of autophagy markers including Beclin-1, ATG5, and LC3 were increased [48]. Another result of this study is the inhibition of the NF-κB signaling pathway after TCA administration [48]. NF-κB, a transcription factor, plays an important role in regulating the release of pro-inflammatory and anti-inflammatory cytokines that are effective in the coordination of the immune system and it induces transcription of pro-inflammatory genes [49].
Another study from Du et al. investigated the anti-inflammatory effect of Maresin 1 (MaR-1) in periodontal ligament cells [50]. Pro-inflammatory cytokine levels in the cells exposed to Porphyromonas gingivalis and its lipopolysaccharide (Pg-LPS) were evaluated after MaR-1 therapy [50]. As known, MaR1 is derived from docosahexaenoic acid and it has an anti-inflammatory effect in many cell types [51]. This study showed decreased levels of pro-inflammatory cytokines such as IL-6, IL-8, TNF-α, and IL-1β after MaR-1 therapy [50]. Also, the activation of autophagy occurs through the glycogen synthase kinase-3β (GSK-3β)/β-catenin signal pathway. As a result of this study, increased LC3I, Beclin-I expression and decreased p62 expression were interpreted as the survival-promoting effect of MaR-1 therapy in periodontal ligament cells [50]. Collectively, these studies demonstrate the efficacy of various chemical components to induce autophagy and maybe a potential therapeutic approach to control the progression of periodontal diseases.
The effect of dental plaque composition on autophagy and periodontal destruction
The dental plaque composition has a significant impact on the growth of periodontal pathogens. Butyrate, which is one of the components of dental plaque is the metabolite of periodontal pathogenic bacteria such as Porphyromonas gingivalis (Pg) [52]. Butyrate inhibits the proliferation of gingival fibroblasts while triggering pro-inflammatory cytokine expression [53]. Tsuda et al. investigated the effect of butyrate on gingival epithelial cells [54]. They found that butyrate induced caspase-independent autophagic cell death and phosphatidylserine redistribution in gingival cells [54]. Also increased LC3 II accumulation (a marker of autophagy) was observed [54]. These results showed that; although autophagy is a programmed cell survival mechanism in both physiologic and pathologic conditions, excessive autophagic activation leads to cell death [55-57].
Reactive oxygen species (ROS) and total antioxidant concentrations In periodontitis and their potential linkage with autophagy
Previously, it was reported that periodontal pathogens alone are not sufficient for the development of tissue damage [58]. Inadequate host immune response and ROS are other accompanying factors for the development of periodontal cell damage [58]. ROS are many chemically reactive molecules derived from molecular oxygen [59]. They play an important role in the regulation of cellular processes. At low concentrations, ROS stimulates the proliferation and differentiation of human periodontal ligament fibroblasts, while at higher concentrations, they have cytotoxic effects on periodontal tissues and participate in pathogen killing [60,61]. Polymorphonuclear leukocytes (PMNLs), osteoclast, fibroblasts, activated in inflammation and they are endogenous sources leading to ROS production [62]. It was found that intracellular production and extracellular secretion of ROS can be induced by stimulation of PMNLs by periodontal pathogens [63]. These produced free radicals evoke bone destruction through NF-kB [64]. They play an important role in the remodeling of alveolar bone affected by periodontitis [65]. Bullon et al. found that peripheral blood mononuclear cells from patients with periodontitis have mitochondrial dysfunction, decreased CoQ10 levels and citrate synthase activity with high levels of ROS production [47]. For the survival of the cell, there are two mechanisms to remove these harmful radicals. The first is the enzymatic antioxidant system, which consists of superoxide dismutase (SOD)/catalase (CAT)/glutathione peroxidase (GPx), neutralize the potentially toxic ROS. Second, the nonenzymatic antioxidant system, which consists of reduced glutathione (GSH), α-tocopherol, ascorbic acid, and coenzyme Q [66].
The intimate relationship between ROS/antioxidant system and periodontitis has long been evaluated. Many recent studies have reported that serum total antioxidant concentrations have been downregulated in patients with periodontitis [67,68]. Concordant results were obtained with previous findings in the study of Brock et al [69]. In this study, antioxidant concentration in gingival crevicular fluid (GCF) was compared between healthy controls and patients with periodontitis. They found that GCF antioxidant concentration was significantly lower in patients with periodontitis compared to healthy controls [69].
Hagio-Izaki et al. showed that intracellular ROS increased in the Porphyromonas gingivalis Lipopolysaccharide (PgLPS) exposed cells [3]. Lipopolysaccharide (LPS), a biologically active endotoxin derived from the cell wall of the gram-negative bacteria, plays a significant role in the pathogenesis of periodontitis [70]. In this study, the upregulation of AMPK activity and autophagy was observed in PgLPS-exposed cells. However, the expression of Beclin-I, LC3, and ROS was suppressed in the cells after N-acetylcysteine (NAC) therapy [3]. NAC, which has anti-ROS activity, is efficient for the ROS suppression in PgLPS-exposed cells. As a result, it is accepted that the deterioration of the homeostatic balance between ROS and enzymatic/nonenzymatic antioxidant systems is an important factor in the development of periodontitis [67].
ROS and nuclear factor erythroid 2-related factor 2(Nrf-2) In periodontitis
Nrf-2 is a transcription activator which has vital importance on cell survival from oxidative stress and inflammation [71,72]. Normally, Nrf-2 localized in the cytoplasm and linked to the protein called Keap1 [73]. When exposed to ROS, Nrf-2 breaks off the Keap1 link and changes position towards the nucleus to increase the expression of genes with Antioxidant Response Elements (ARE) [72,73]. The association of Nrf-2 with AREs in the nucleus initiates the transcription of several antioxidant genes [74,75] (Figure 3).
To elucidate whether Nrf-2 increased the genomic expression of antioxidant enzymes, Sima et al. investigated the oxidative state of polymorphonuclear neutrophils in patients with periodontitis [76]. They found that Nrf-2expression has been downregulated in periodontitis and this leads to severe bone loss associated with high oxidative stress [76]. It has also been shown that osteoclastic bone resorption is substantially promoted in Nrf-2 deficient osteoclast precursor cells [77]. For the moment, suffice it to say that Nrf-2 has significant importance in oxidative-stress-associated periodontal damage.
Hypoxia and autophagy
Hypoxia is common environmental stress involved in many pathophysiological conditions [78,79]. The activation of angiogenesis, osteoclast maturation, and anaerobic glycolysis are regulated by hypoxia [80,81]. Several recent studies have shown that hypoxia could promote osteoclast differentiation [82-84]. Osteoclasts, which have been demonstrated as monocyte-macrophage derived multinuclear cells, are principal resorptive cells of bone [85]. Arnett et al. showed that hypoxia stimulates bone resorption by accelerating the formation of large osteoclasts [82]. Besides, Chung et al. showed that the Beclin-1 plays an important role in receptor activator of nuclear factor-kB (NF-kB) ligand (RANKL) induced osteoclast differentiation by inducing the production of reactive oxygen species [86].
As an essential regulator in hypoxia-induced bone development, Hypoxia-induced factor-1 alpha (HIF-1α) has an important role in chondrogenesis and osteogenesis [87]. Huang et al. investigated HIF-1α, BNIP3, pAMPK, and autophagy-related proteins (LC3, beclin-1, Atg5–12, and p62) in human inflammatory periapical lesions [88]. They evaluated samples from patients with radicular cysts (RCs), Periapical Granulomas (PGs) and healthy dental pulp tissues. After immunohistochemical analysis, the expression of autophagy and hypoxia-related proteins in each group were compared with each other [88]. In periapical lesions, HIF-1α, Bnip3, pAMPK, LC3II, and Atg5–12 proteins were significantly higher than those in healthy dental pulp tissues [88]. The results showed that autophagy is involved in the development and maintenance of hypoxia-associated periapical lesions.
It has long been recognized that hypoxia increases the expression of several cytokines in human Periodontal Ligament Cells (PDLCs), gingival fibroblasts and synovial fibroblasts [89,90]. Song et. al investigated the effects of hypoxia on human Periodontal Ligament Cells (PDLCs) [91]. They used CoCl2 to establish hypoxic conditions. Results indicated that increased expression of, IL-1β, MMP-8, Bnip3 mRNA and LC3-I in CoCl2 treated cells [91]. However, these values were reversed in cells treated with N-acetylcysteine (NAC). This evidence suggests that CoCl2 induced hypoxia results in hypoxic cell death in human PDLCs via HIF-1α pathway [91].
Intriguingly, immunohistochemical analysis showed that HIF-1α protein was present in detectable amounts in benign tumors, increased amounts in malignant tumors, and excessive amounts in metastases, although not in normal tissues [92,93]. This overexpression of HIF has been possible as a result of intratumoral hypoxia or genetic alterations [94,95]. As the tumor size increases, the tumor mass gradually becomes hypoxic until sufficient blood vessels are formed. So, the importance of hypoxia in the cellular process through gene expression and its critical role in oxygen hemostasis suggest that it acts as the conductor of cell metabolic function.
Role of autophagy in oral cancer
Oral cancers are one of the major causes of morbidity and mortality, including a wide range of cancers originating from mucosal, salivary gland, or other soft and hard tissues of the mouth. More than 90% of these are squamous cell carcinomas that develop from the surface epithelium of the mucosa [96,97]. Genetic predisposition and exposure to environmental carcinogens such as tobacco, alcohol, chronic inflammation, and viral infection are the major etiologic factors for Oral Squamous Cell Carcinomas (OSCC) development [98]. Sambandam et al. investigated the autophagosome marker proteins, LC3-II and ATG5 expression in OSCC specimens [99]. They identified high levels of LC3-II in these tumor cells. Similar to this study, high levels of several autophagy-related genes (Atg), Beclin 1 and LC3-I have been detected in gastrointestinal cancers [100,101]. Also, autophagosome formation was detected in invasive and metastatic melanoma cells [102].
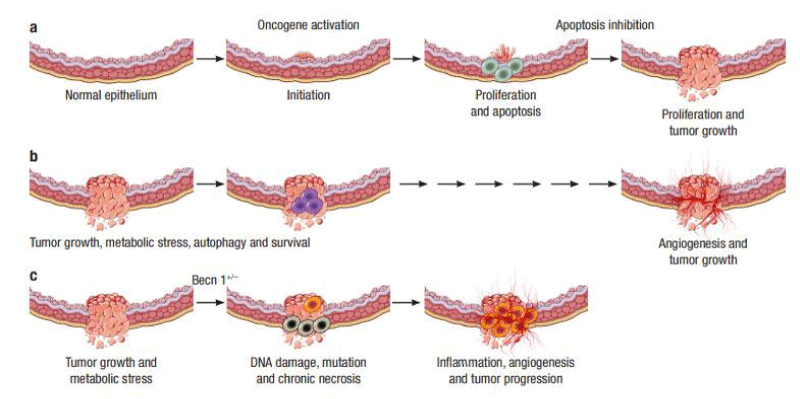
Autophagy is a highly evolutionarily conservative cellular metabolic pathway. Accumulating pieces of evidence suggest that autophagy is a switchable mechanism in cancer progression [103,104]. Autophagy can inhibit the initial stages of carcinogenesis [105], but also support the survival and growth of established cancers [106-108]. Yue et al. investigated the role of Beclin 1 and autophagy in tissue homeostasis [109]. They found that the deletion of Beclin 1 in mice contributes to tumor formation through the deregulation of autophagy [109]. On the other hand, Degenhardt et al. showed that defects in apoptosis due to either Bax/Bak deficiency or Bcl-2 expression reveal autophagy in response to metabolic stress, which is required for cell survival [110]. The growth of tumor cells is initially limited due to insufficient blood supply. Then, autophagy and angiogenesis are triggered to withstand this hypoxic environment, resulting in the survival of tumor cells [110,111] (Figure 4).
Recent evidence suggests that autophagy provides a protective function to limit tumor necrosis and inflammation, and to mitigate genome damage in tumor cells in response to metabolic stress (Figure 4) [111]. However, in tumor cells possessing apoptotic and autophagy defects, necrotic cell death is triggered in metabolically stressed tumor regions. This necrosis develops due to the activation of an inflammatory response with the recruitment of inflammatory cells, cytokine production, and nuclear factor-κB (NFκB) activation and followed by DNA damage and tumor progression [110,112-115].
The mechanisms that regulate the mutually opposed survival-supporting and death-promoting roles for autophagy are still far from resolution. The most plausible explanation is that catabolism through autophagy is predominantly survival-supporting, but that an imbalance in cell metabolism, where autophagic cellular consumption exceeds the cellular capacity for synthesis, promotes cell death. Although current studies address the role of autophagy in tumor cells, there is a need to investigate how autophagy can be controlled and regulated in suppressing tumor growth.
Conclusion
Understanding the autophagy mechanism and their interrelationships with oral diseases will lead the way in the discovery of novel treatment modalities. This review highlights the importance of the autophagy mechanism in the maintenance of oral tissue health. Future studies will likely be focused on understanding the association with autophagy and oral diseases.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
- Yoshimori T (2004) Autophagy: A regulated bulk degradation process inside cells. Biochem Biophys Res Commun 313: 453-458. Link: https://bit.ly/2Utprxi
- Mizushima N, Yoshimori T, Levine B (2010) Methods in mammalian autophagy research. Cell 140: 313-326. Link: https://bit.ly/33Zx6qc
- Hagio-Izaki K, Yasunaga M, Yamaguchi M, Kajiya H, Morita H, et al. (2018) Lipopolysaccharide induces bacterial autophagy in epithelial keratinocytes of the gingival sulcus. BMC Cell Biol 19: 18. Link: https://bit.ly/2Jv7eZS
- Singh R, Cuervo AM (2011) Autophagy in the cellular energetic balance. Cell Metab 13: 495-504. Link: https://bit.ly/2RdVM9F
- Yang YP, Liang ZQ, Gu ZL, Qin ZH (2005) Molecular mechanism and regulation of autophagy. Acta Pharmacol Sin 26: 1421-1434. Link: https://bit.ly/3bxyfYK
- Kaushik S, Cuervo AM (2018) The coming of age of chaperone-mediated autophagy. Nat Rev Mol Cell Biol 19: 365-381. Link: https://bit.ly/33W7hHI
- Massey AC, Zhang C, Cuervo AM (2006) Chaperone‐mediated autophagy in aging and disease. Curr Top Dev Biol 73: 205-235. Link: https://bit.ly/2UwcrHt
- Kon M, Cuervo AM (2010) Chaperone‐mediated autophagy in health and disease. FEBS Lett 584: 1399-1404. Link: https://bit.ly/2QXpbEy
- Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, et al. (2017) Molecular definitions of autophagy and related processes. Embo J 36: 1811-1836. Link: https://bit.ly/2yjDv3T
- Li WW, Li J, Bao JK (2012) Microautophagy: lesser-known self-eating. Cell Mol Life Sci 69: 1125-1136. Link: https://bit.ly/2Ut1DK7
- Mizushima N, Komatsu M (2011) Autophagy: Renovation of cells and tissues. Cell 147: 728-741. Link: https://bit.ly/2w5ZsCY
- Tan YQ, Zhang J, Zhou G (2017) Autophagy and its implication in human oral diseases. Autophagy 13: 225-236. Link: https://bit.ly/2UPtMtS
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, et al. (2011) Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331: 456-461. Link: https://bit.ly/3dK6CgX
- Kim J, Kim YC, Fang C, Russell RC, Kim JH, et al. (2013) Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell 152: 290-303. Link: https://bit.ly/3awVH8u
- Russell RC, Tian Y, Yuan H, Park HW, Chang YY, et al. (2013) ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 15: 741-750. Link: https://bit.ly/3dI18U4
- Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, et al. (2000) The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol 151: 263-276. Link: https://bit.ly/39sT0na
- Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, et al. (2001) Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol 152: 657-668. Link: https://bit.ly/3bGbyBA
- Anding AL, Baehrecke EH (2015) Autophagy in cell life and cell death. Curr Top Dev Biol 114: 67-91. Link: https://bit.ly/2QZ6NeB
- Rubinsztein DC, Cuervo AM, Ravikumar B, Sarkar S, Korolchuk VI, et al. (2009) In search of an ‘‘autophagomometer’’. Autophagy 5: 585–589. Link: https://bit.ly/2QX1ufS
- Wild P, McEwan DG, Dikic I (2014) The LC3 interactome at a glance. J Cell Sci 127: 3-9. Link: https://bit.ly/3bCy6TV
- Fujita N, Hayashi-Nishino M, Fukumoto H, Omori H, Yamamoto A, et al. (2008) An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell 19: 4651-4659. Link: https://bit.ly/2UOh5PV
- Yu L, Chen Y, Tooze SA (2018) Autophagy pathway: Cellular and molecular mechanisms. Autophagy 14: 207-215. Link: https://bit.ly/341Xa47
- Huang Y, Mucke L (2012) Alzheimer mechanisms and therapeutic strategies. Cell 148: 1204-1222. Link: https://bit.ly/2Jqs6BR
- Kenific CM, Debnath J (2015) Cellular and metabolic functions for autophagy in cancer cells. Trends Cell Biol 25: 37-45. Link: https://bit.ly/2QYO3vR
- Netea-Maier RT, Plantinga TS, van de Veerdonk FL, Smit JW, Netea MG (2016) Modulation of inflammation by autophagy: consequences for human disease. Autophagy 12: 245-260. Link: https://bit.ly/2UslWaq
- Mizushima N (2005) The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ 12: 1535-1541. Link: https://go.nature.com/2ULKytK
- Thomas N, Choi HK, Wei X, Wang L, Mishina Y, et al. (2019) Autophagy Regulates Craniofacial Bone Acquisition. Calcified Tissue Int 105: 518-530. Link: https://bit.ly/3dH6xLe
- Wei X, Thomas N, Hatch NE, Hu M, Liu F (2017) Postnatal craniofacial skeletal development of female C57BL/6NCrl mice. Front Physiol 8: 697. Link: https://bit.ly/2WUZxEv
- Amizuka N, Davidson D, Liu H, Valverde- Franco G, Chai S, et al. (2004) Signalling by fibroblast growth factor receptor 3 and parathyroid hormone-related peptide coordinate cartilage and bone development. Bone 34: 13-25. Link: https://bit.ly/3dGml0t
- Stevens DA, Hasserjian RP, Robson H, Siebler T, Shalet SM (2000) Thyroid hormones regulate hypertrophic chondrocyte differentiation and expression of parathyroid hormone-related peptide and its receptor during endochondral bone formation. J Bone Miner Res 15: 2431-2442. Link: https://bit.ly/2USmsgU
- Onal M, Piemontese M, Xiong J, Wang Y, Han L, et al. (2013) Suppression of autophagy in osteocytes mimics skeletal aging. J Biol Chem 288:17432-17440. Link: https://bit.ly/2QZXLho
- Harada SI, Rodan GA (2003) Control of osteoblast function and regulation of bone mass. Nature 423: 349-355. Link: https://go.nature.com/2Uu2cmK
- Nollet M, Santucci-Darmanin S, Breuil V, Al-Sahlanee R, Cros C, et al. (2014) Autophagy in osteoblasts is involved in mineralization and bone homeostasis. Autophagy 10: 1965-1977. Link: https://bit.ly/2wSrIsS
- Al-Barghouthi BM, Farber CR (2018) Dissecting the genetics of osteoporosis using systems approaches. Trends Genet 35: 55-67. Link: https://bit.ly/2UuN5K1
- Li H, Li D, Ma Z, Qian Z, Kang X, et al. (2018) Defective autophagy in osteoblasts induces endoplasmic reticulum stress and causes remarkable bone loss. Autophagy 14: 1726-1741. Link: https://bit.ly/2QYHcm0
- Hamrick I, Schrager S, Nye AM (2015) Treatment of osteoporosis: Current state of the art. Wien Med Wochenschr 165: 54-64. Link: https://bit.ly/341XYpF
- Whittier X, Saag KG (2016) Glucocorticoid-induced osteoporosis. Rheum Dis Clin North Am 42:177-189. Link: https://bit.ly/2R01yeJ
- Shapiro IM, Layfield R, Lotz M, Settembre C, Whitehouse C (2014) Boning up on autophagy: the role of autophagy in skeletal biology. Autophagy 10: 7-19. Link: https://bit.ly/2w12bgV
- Demmer RT, Papapanou PN (2010) Epidemiologic patterns of chronic and aggressive periodontitis. Periodontol 2000 53: 28-44. Link: https://bit.ly/2xEr0iV
- Pihlstrom BC, Bryan SM, Johnson NW (2005) Periodontal Disease. Lancet 366: 1809-1820. Link: https://bit.ly/2WXPFJV
- Offenbacher S (1996) Periodontal disease: Pathogenesis. Ann Periodontol 1: 821-878. Link: https://bit.ly/2JtMB0u
- Van Dyke TE, Dave S (2005) Risk factors for periodontitis. J Int Acad Periodontol 7: 3-7. Link: https://bit.ly/2w61gvK
- Griffen AL, Becker MR, Lyons SR, Moeschberger ML, Leys EJ (1998) Prevalence of Porphyromonas gingivalis and periodontal health status. J Clin Microbiol 36: 3239-3242. Link: https://bit.ly/2V20qsr
- Lang NP, Löe H (1972) The relationship between the width of keratinized gingiva and gingival health. J Periodontol 43: 623-627. Link: https://bit.ly/39xcYNk
- Berglundh T, Zitzmann NU, Donati M (2011) Are peri‐implantitis lesions different from periodontitis lesions? J Clin Periodontol 38: 188-202. Link: https://bit.ly/2USZarc
- Newman MG, Takei HH, Carranza FA (2006) Carranza’s Clinical Periodontology, 10 ed. Saunders Company 506- 512.
- Bullon P, Cordero MD, Quiles JL, del Carmen Ramirez-Tortosa M, Gonzalez-Alonso A, et al. (2012) Autophagy in periodontitis patients and gingival fibroblasts: unraveling the link between chronic diseases and inflammation. BMC Med 10: 122. Link: https://bit.ly/2QWqPXl
- Chung J, Kim S, Lee HA, Park MH, Kim S, et al. (2018) Trans‐cinnamicaldehyde inhibits Aggregatibacter actinomycetemcomitans‐induced inflammation in THP‐1–derived macrophages via autophagy activation. J Periodontol 89: 1262-1271. Link: https://bit.ly/2UvWuRG
- Arabaci T, Cicek Y, Canakci V, Canakcı CF, Ozgoz M, et al. (2010) Immunohistochemical and Stereologic Analysis of NF-kappaB Activation in Chronic Periodontitis. Eur J Dent 4: 454-461. Link: https://bit.ly/3auOntT
- Du L, Li Y, Liu W (2018) Maresin 1 regulates autophagy and inflammation in human periodontal ligament cells through glycogen synthase kinase–3β/β-catenin pathway under inflammatory conditions. Arch Oral Biol 87: 242-247. Link: https://bit.ly/2URaiVt
- Serhan CN, Yang R, Martinod K, Kasuga K, Pillai PS, et al. (2009) Maresins: Novel macrophage mediators with potent antiinflammatory and proresolving actions. J Exp Med 206: 15-23. Link: https://bit.ly/2UvwvJW
- Pöllänen MT, Salonen JI, Uitto VJ (2003) Structure and function of the tooth-epithelial interface in health and disease. Periodontol 2000 31: 12-31. Link: https://bit.ly/3awsJFF
- Jeng JH, Chan CP, Ho YS, Lan WH, Hsieh CC, Chang MC (1999) Effects of butyrate and propionate on the adhesion, growth, cell cycle kinetics, and protein synthesis of cultured human gingival fibroblasts. J Periodontol 70:1435-1442. Link: https://bit.ly/2UwwVj2
- Tsuda H, Ochiai K, Suzuki N, Otsuka K (2010) Butyrate, a bacterial metabolite, induces apoptosis and autophagic cell death in gingival epithelial cells. J Periodontal Res 45: 626-634. Link: https://bit.ly/3bB38vq
- González-Polo RA, Boya P, Pauleau AL, Jalil A, Larochette N, et al. (2005) The apoptosis/autophagy paradox: autophagic vacuolization before apoptotic death. J Cell Sci 118: 3091-3102. Link: https://bit.ly/2UOjeer
- Liu JL, Chen FF, Lung J, Lo CH, Lee FH, et al. (2014) Prognostic significance of p62/SQSTM1 subcellular localization and LC3B in oral squamous cell carcinoma. Br J Cancer 111: 944-954. Link: https://bit.ly/2UKes1w
- Levine B, Yuan J (2005) Autophagy in cell death: An innocent convict? J Clin Invest 115: 2679-2688. Link: https://bit.ly/2JsoDmk
- Hajishengallis G (2015) Periodontitis: From microbial immune subversion to systemic inflammation. Nat Rev Immunol 15: 30-44. Link: https://bit.ly/3awk8Tj
- DiMeo S, Reed TT, Venditti P, Victor VM (2016) Role of ROS and RNS sources in physiological and pathological conditions. Oxid Med Cell Longev. Link: https://bit.ly/2w59WCy
- Saita M, Kaneko J, Sato T, Takahashi SS, Wada-Takahshi S, et al. (2016) Novel antioxidative nanotherapeutics in a rat periodontitis model: reactive oxygen species scavenging by redox injectable gel suppresses alveolar bone resorption. Biomaterials 76: 292-301. Link: https://bit.ly/2URN4yL
- Galli C, Passeri G, Macaluso GM (2011) FoxOs, Wnts and oxidative stress-induced bone loss: new players in the periodontitis arena? J Periodont Res 46: 397-406. Link: https://bit.ly/2UuspSf
- Çanakçı CF, Tatar A, Çanakçı V, Cicek Y, Oztas S, et al. (2006) New evidence of premature oxidative DNA damage: mitochondrial DNA deletion in gingival tissue of patients with periodontitis. J Periodontol 77: 1894-1900. Link: https://bit.ly/2JqwJvJ
- Aoshiba K, Nagai A (2003) Oxidative stress, cell death, and other damage to alveolar epithelial cells induced by cigarette smoke. Tabacco Induc Dis 1: 219-226. Link: https://bit.ly/2JoXrVd
- Gan P, Gao Z, Zhao X, Qi G (2016) Surfactin inducing mitochondria dependent ROS to activate MAPKs, NF-kappaB and inflammasomes in macrophages for adjuvant activity. Sci Rep 6: 39303. Link: https://go.nature.com/2UHHte4
- Çanakçı CF, Çiçek Y, Çanakçı V (2005) Reactive oxygen species and human inflammatory periodontal diseases. Biocem (Moscow) 70: 619-628. Link: https://bit.ly/2JpXn7N
- He L, He T, Farrar S, Ji L, Liu T, et al. (2017) Antioxidants maintain cellular redox homeostasis by elimination of reactive oxygen species. Cell Physiol Biochem 44: 532-553. Link: https://bit.ly/2JodO4D
- Patil VS, Patil VP, Gokhale N, Acharya A, Kangokar P (2016) Chronic periodontitis in type 2 diabetes mellitus: oxidative stress as a common factor in periodontal tissue injury. J Clin Diagn Res 10: 12-16. Link: https://bit.ly/3aw9Pil
- Thomas B, Madani SM, Prasad BR, Kumari S (2014) Comparative evaluation of serum antioxidant levels in periodontally diseased patients: An interventional study. Contemp Clin Dent 5: 340-344. Link: https://bit.ly/3dHrNQP
- Brock GR, Butterworth CJ, Matthews JB, Chapple ILC (2004) Local and systemic total antioxidant capacity in periodontitis and health. J Clin Periodontol 31: 515-521. Link: https://bit.ly/2QXGsO6
- Xu YI, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE (2007) Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity 27: 135-144. Link: https://bit.ly/3bBvENw
- Yu X, Kensler T (2005) Nrf2 as a target for cancer chemoprevention. Mutat Res 591: 93-102. Link: https://bit.ly/2WVQ5k1
- Niture SK, Khatri R, Jaiswal AK (2014) Regulation of Nrf2-an update. Free Radic Biol Med 66: 36-44. Link: https://bit.ly/3bPo6qL
- Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T (2006) Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol Cell Biol 26: 2887-2900. Link: https://bit.ly/3ayqIc7
- Ma Q (2013) Role of nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol 53: 401-426. Link: https://bit.ly/2QUqcNO
- Liu C, Mo L, Niu Y, Li X, Zhou X (2017) The role of reactive oxygen species and autophagy in periodontitis and their potential linkage. Front Physiol 8: 439. Link: https://bit.ly/2UJednq
- Sima C, Aboodi GM, Lakschevitz FS, Sun C, Goldberg MB (2016) Nuclear factor erythroid 2-related factor 2 down-regulation in oral neutrophils is associated with periodontal oxidative damage and severe chronic periodontitis. Am J Pathol 186: 1417-1426. Link: https://bit.ly/2xwSF5v
- Hyeon S, Lee H, Yang Y, Jeong W (2013) Nrf2 deficiency induces oxidative stress and promotes RANKL-induced osteoclast differentiation. Free Radic Biol Med 65: 789-799. Link: https://bit.ly/341S0W2
- Gaber T, Dziurla R, Tripmacher R, Burmester GR, Buttgereit F (2005) Hypoxia inducible factor (HIF) in rheumatology: Low O2! See what HIF can do. Ann Rheum Dis 64: 971-980. Link: https://bit.ly/2WUIu5j
- Watanabe Y, Terashima Y, Takenaka N, Kobayashi M, Matsushita T (2007) Prediction of avascular necrosis of the femoral head by measuring intramedullary oxygen tension after femoral neck fracture. J Orthop Trauma 21: 456-461. Link: https://bit.ly/3axAZFr
- Avni R, Cohen B, Neeman M (2011) Hypoxic stres and cancer: Imaging the axis of evilin tumor metastasis. NMR Biomed 24: 569-581. Link: https://bit.ly/2Uu7bnt
- Bahadori B, Uitz E, Mayer A, Harauer J, Dam K, et al. (2010) Polymorphisms of the hypoxia-inducible factor 1 gene peripheral artery disease. Vasc Med 15: 371-374. Link: https://bit.ly/2JpYrbN
- Arnett TR, Gibbons DC, Utting JC, Orriss IR, Hoebertz A, et al. (2003) Hypoxia is a major stimulator of osteoclast formation and bone resorption. J Cell Physiol 196: 2-8. Link: https://bit.ly/39qKkxn
- Srinivasan S, Avadhani NG (2007) Hypoxia-mediated mitochondrial stress in RAW264.7cells induces osteoclast-like TRAP-positive cells. Ann NY Acad Sci 1117: 51–61. Link: https://bit.ly/2xBgo4u
- Knowles HJ, Athanasou NA (2009) Acute hypoxia and osteoclast activity: A balance between enhanced resorption and increased apoptosis. J Pathol 218: 256-264. Link: https://bit.ly/2Uw0CRG
- Maziere C, Louvet L, Gomila C, Kamel S, Massy Z (2009) Oxidized low density lipoprotein decreases Rankl-induced differentiation of osteoclasts by inhibition of Rankl signaling. J Cell Physiol 221: 572–578. Link: https://bit.ly/2X2TFsC
- Chung YH, Jang Y, Choi B, Song DH, Lee EJ, et al. (2014) Beclin‐1 is required for RANKL‐induced osteoclast differentiation. J Cell Physiol 229: 1963-1971. Link: https://bit.ly/2R07aFP
- Araldi E, Schipani E (2010) Hypoxia, HIFs and bone development. Bone 47: 190-196. Link: https://bit.ly/3dAAqwu
- Huang HY, Wang WC, Lin PY, Huang CP, Chen CY, et al. (2018) The roles of autophagy and hypoxia in human inflammatory periapical lesions. Int Endod J 51: 125-145. Link: https://bit.ly/2wHJnnk
- Motohira H, Hayashi J, Tatsumi J, Tajima M, Sakagami H, et al. (2007) Hypoxia and reoxygenation augment bone-resorbing factor production from human periodontal ligament cells. J Periodontol 78: 1803-1809. Link: https://bit.ly/2wOPLsP
- Thornton RD, Lane P, Borghaei RC, Pease EA, Caro J, et al. (2000) Interleukin 1 induces hypoxia-inducible factor 1 in human gingival and synovial fibroblasts. Biochem J 350: 307-312. Link: https://bit.ly/2xBgn0B
- Song ZC, Zhou W, Shu R, Ni J (2012) Hypoxia induces apoptosis and autophagic cell death in human periodontal ligament cells through HIF‐1α pathway. Cell Prolif 45: 239-248. Link: https://bit.ly/39x3t0T
- Stiehl DP, Jelkmann W, Wenger RH, Hellwig- Bürgel T (2002) Normoxic induction of the hypoxia-inducible factor 1 alpha by insulin and interleukin-1 beta involves the phosphatidylinositol 3-kinase pathway. FEBS Lett 512: 157-162. Link: https://bit.ly/3dG4yXt
- Tian H, McKnight SL, Russell DW (1997) Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev 11: 72-82. Link: https://bit.ly/3dJwhX1
- Brusselmans K, Bono F, Maxwell P, Dor Y, Dewerchin M, et al. (2001) Hypoxia-inducible factor 2-α (HIF-2) is involved in the apoptotic response to hypoglycemia but not to hypoxia. J Biol Chem 276: 39192-39196. Link: https://bit.ly/2xDCfbq
- Sodhi A, Montaner S, Patel V, Zohar M, Bais C, et al. (2000) The Kaposi’s sarcoma-associated herpes virus G protein-coupled receptor upregulates vascular endothelial growth factor expression and secretion through mitogen-activated protein kinase and p38 pathways acting on hypoxia-inducible factor 1alpha. Cancer Res 60: 4873-4880. Link: https://bit.ly/3auugfq
- Regezi JA, Schiubba JJ (1993) Oral Pathology. Clinical- Pathological Correlations. 2nd ed. Philadelphia, W.B. Saunders Company. Link: https://bit.ly/2QZh9eC
- Cawson RH, Odell EW (1997) Essentials of Oral Pathology and Oral Medicine, 6th ed. London, Churchill Livingstone.
- Choi S, Myers JN (2008) Molecular pathogenesis of oral squamous cell carcinoma: implications for therapy. J Dent Res 87: 14-32. Link: https://bit.ly/2QXAGfc
- Sambandam Y, Sakamuri S, Balasubramanian S, Haque A (2016) RANK ligand modulation of autophagy in oral squamous cell carcinoma tumor cells. J Cell Biochem 117: 118-125. Link: https://bit.ly/39xlMTo
- Ahn CH, Jeong EG, Lee JW, Kim MS, Kim SH, et al. (2007) Expression of beclin‐1, an autophagy‐related protein, in gastric and colorectal cancers. Apmis 115: 1344-1349. Link: https://bit.ly/3bCTDfb
- Yoshioka A, Miyata H, Doki Y, Yamasaki M, Sohma I, et al. (2008) LC3, an autophagosome marker, is highly expressed in gastrointestinal cancers. Int. J Oncol 33: 461-468. Link: https://bit.ly/2UvUh8M
- Lazova R, Klump V, Pawelek J (2010) Autophagy in cutaneous malignant melanoma. J Cutan Pathol 37: 256-268. Link: https://bit.ly/3bEU5tC
- Amaravadi R, Kimmelman AC, White E (2016) Recent insights into the function of autophagy in cancer. Genes Dev 30: 1913-1930. Link: https://bit.ly/2UyF5YB
- Kimmelman AC, White E (2017) Autophagy and tumor metabolism. Cell Metab 25: 1037-1043. Link: https://bit.ly/3bIcNjJ
- Muhammad JS, Nanjo S, Ando T, Yamashita S, Maekita T, et al. (2017) Autophagy impairment by Helicobacter pylori‐induced methylation silencing of MAP1LC3Av1 promotes gastric carcinogenesis. Int J Cancer 140: 2272-2283. Link: https://bit.ly/2w1nFdr
- Shen S, Zhou M, Huang K, Wu Y, Ma Y, et al. (2017) Blocking autophagy enhances the apoptotic effect of 18β-glycyrrhetinic acid on human sarcoma cells via endoplasmic reticulum stress and JNK activation. Cell Death Dis 8: 3055. Link: https://bit.ly/3azo1XY
- Kong P, Zhu X, Geng Q, Xia L, Sun X, et al. (2017) The microRNA-423-3p-Bim axis promotes cancer progression and activates oncogenic autophagy in gastric cancer. Mol Ther 25: 1027-1037. Link: https://bit.ly/2URmQfx
- Wang J, Liu Z, Hu T, Han L, Yu S, et al. (2017) Nrf2 promotes progression of non-small cell lung cancer through activating autophagy. Cell Cycle 16: 1053-1062. Link: https://bit.ly/2WXXSho
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N (2003) Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci 100: 15077-15082. Link: https://bit.ly/2QXOBSx
- Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, et al. (2006) Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer cell 10: 51-64. Link: https://bit.ly/2w4CWu8
- Mathew R, Karantza-Wadsworth V, White E (2007) Role of autophagy in cancer. Nat Rev Cancer7: 961-967. Link: https://bit.ly/2wQI7ht
- Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, et al. (2007) Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev 21: 1621-1635. Link: https://bit.ly/2JqZ5pp
- Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, et al. (2007) Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev 21: 1367-1381. Link: https://bit.ly/2wNJWf6
- Karin M (2006) Nuclear factor-κB in cancer development and progression. Nature 441: 431. Link: https://bit.ly/2QZlo9T
- Karin M, Lawrence T, Nizet V (2006) Innate immunity gone awry: linking microbial infections to chronic inflammation and cancer. Cell 124: 823-835. Link: https://bit.ly/3aChb40
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.



 Save to Mendeley
Save to Mendeley
