Journal of Clinical Microbiology and Biochemical Technology
Retroviruses: Reversing the dogma of life - A review
Shobha Potlakayala1*, Andrew Miles2, Shrina Patel1, Aayushi Patel1, Gregory Wolbrette3, Shriya Kane4, Nicole Lookfong1, Abigayle Noble1, Xiuli Shen1 and Sairam Rudrabhatla1
2Penn State University Park, State College, PA, United States of America
3Physician Assistant Studies Program, Shenandoah University, Winchester, VA, United States of America
4University of Iowa Hospitals and Clinics, Iowa City, IA, United States of America
Cite this as
Potlakayala S, Miles A, Patel S, Patel A, Wolbrette G, et al. (2022) Retroviruses: Reversing the dogma of life - A review. J Clin Microbiol Biochem Technol 8(1): 018-028. DOI: 10.17352/jcmbt.000051Copyright
© 2022 Potlakayala S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Retroviruses replicate by means of reverse transcription, utilizing an enzyme, reverse transcriptase, in conjunction with integrase. Their elements have been found in humans, animals, fungi, plants, and bacteria alike. For millions of years, these elements are continuing to integrate into the eukaryotic genomes and affecting these organisms to date. Specifically, endogenous retroviruses have been shown to comprise a large portion of vertebrate genomes. Studies have shown that these microscopic viral elements within the genome are influencing gene expression and in turn evolution, by affecting adjacent gene expression patterns. In the medical field, these retroviruses can present illnesses for many, such as those living with Human Immunodeficiency Virus or Human T Cell Lymphotropic Viruses. With modern advances in bioinformatics, genomics, and drug design, retroviruses are being understood much better. A multitude of new discoveries is advancing the scientific communities to mitigate, prevent, and hopefully cure serious medical ailments caused by retroviruses.
Abbreviations
AIDS: Acquired Immunodeficiency Syndrome; ART: Antiretroviral Therapy; Cas: CRISPR-associated; C-C CCR5: chemokine Receptor Type 5; cDNA: complementary DNA; CRISPR: Clustered Regularly Interspaced Short Palindromic Repeats; CXCR4: C-X-C Chemokine Receptor 4; dsDNA: double-stranded DNA; dsRNA: double-stranded RNA; ERVs: Endogenous Retroviruses; FVs: Foamy Viruses; HAM/TSP: HTLV-1–Associated Myelopathy/Tropical Spastic Paraparesis; HDR: Homology Directed Repair; HERVs: Human Endogenous Retroviruses; HIV: Human Immunodeficiency Virus; HSCT: Hematopoietic Stem Cell Transplantation; HTLV: Human T-Cell Leukemia Virus; HTLV-1: Human T-Cell Leukemia Virus Type 1; HTLV-2: Human T-Cell Leukemia Virus Type 2; IAP1: Intracisternal A-Particles Class 1; LINEs: Long Interspersed Repetitive Elements; LTR-RTs: Long Terminal Repeats-Retrotransposons; LTRs: Long Terminal Repeats; NHEJ: Non-Homologous End Joining Repair; NPCs: Neuronal Progenitor Cells; PAM: Photospacer Adjacent Motif; PBS: Primer-Binding Sequence; PDCD-1: Programmed Cell Death Protein 1; PMFV: Permanent Human Fibroblast Virus; sgRNA: single-guide RNA; SINEs: Short Interspersed Repetitive Elements; ssDNA: single-stranded DNA; ssRNA: single-stranded RNA; STLVs: Simian T-Lymphotropic Viruses; TALENs: Transcription Activator-Like Effector Nucleases; TEs: Transposable Elements; TRAC: T-Cell Receptor α Chain; TRBC: T-cell Receptor β Chain; TRIM28: Tripartite Motif-Containing Protein 28; ZFN: Zinc-Finger Nuclease
Introduction
The Central Dogma of Molecular Biology – it possesses an immutable, steadfast quality, its name donating an intrinsic law of permanence. Yet, with the discovery of one simple, infinitely small particle, this indisputable, fundamental biological principle was slightly flawed in its original context. The flow of genetic information, as detailed in any course or textbook beginning with the Central Dogma, always began with DNA, then transcribed to RNA, and, finally, ended with a completely folded protein [1]. However, since the discovery of viruses and retroviruses, modifications and amendments to the Central Dogma were required, as the genetic flow of information then possessed a quality of reversibility- acquired through the study of retroviruses [2]. Because of their unique genome replication process, the study of these retroviruses has had a considerable impact on the diverse field of biology ranging from biotechnology, microbiology, molecular genetics, medicine, and many others. The study of retroviruses also is crucial in the aims of understanding evolution, as a large part of the mammalian genomes seem to be directly from the process of reverse transcription [2]. Having direct links to evolutionary processes as well as diseases brings about the pertinence of advancing insight into retrovirus behavior, genome replication, and how they can positively or negatively impact the day-to-day lives of humans.
These viruses, through their integration into genomes, increase genome diversity within the biosphere [3]. The functionality of viruses on the genomes of bacteria and archaea compared to eukaryotic plant and animal viromes differ, as the largest difference is that for prokaryotes the virome consists largely of double-stranded DNA viruses, while eukaryotes have genomes that are made up of RNA or retrovirus elements [2,4]. Due to the documentation of these retroviruses being coupled with eukaryotes from early in their evolutionary beginnings, it is possible that they coevolved together. One evolutionary example is the development of cytosol as its environmental conditions of it are well-made for retrovirus replication [5]. In terms of the history of viruses, it seems that a single gene is not conserved across all known viruses. They have likely evolved more than once in origin. Even though not a single specific gene is shared among all viruses, they do have genes that correspond to different groupings and families of viruses, leading to the ability to at least classify them accordingly [6].
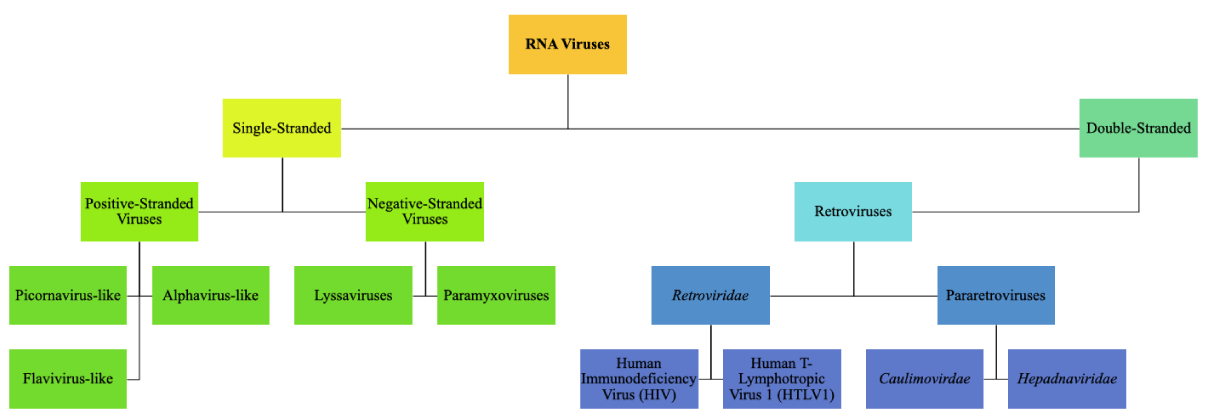
The viruses that are at interplay with RNA are divided into three main categorical groupings: positive and negative-stranded viruses and double-stranded viruses. Similar to all of them is the ability to synthesize RNA-dependent RNA polymerases. These polymerases have a wide range of functions for viruses such as replication and transcription [4]. The most abundant RNA viruses are the positive-stand viruses, and they are very diverse. This class is divided into three main groupings: picornavirus-like, alphavirus-like, and flavivirus-like [7]. Due to the nature of the picornavirus and alphavirus having well-shown partially conserved regions between the two subfamilies, monophyly is hypothesized. Furthermore, the flavivirus-like group remains undetermined in origin at this point as it contains plant and animal viruses that process genomes that are dissimilar [4]. Retroviruses can be divided into two different types: pararetroviruses and Retroviridae. Pararetroviruses have circular, double-stranded DNA and include two types: Caulimoviridae which infect plants, and Hepadnaviridae which infect animals. Viruses belonging to the Retroviridae type possess positive-stranded RNA, and using the enzyme reverse transcriptase, they create complementary DNA (cDNA) to enter the host’s DNA and infect only vertebrate hosts [4,8] Figure 1.
This review seeks to address the evolution, structure, and replication mechanisms of these viruses as well as specifically looking at the bacterial, plant, and human retroviruses. In addition, the review showcases applications and future research, with a primary emphasis on humans and animals.
Viruses
Viruses are microbes that contain DNA or RNA as their genetic material enclosed by a protein coat. Viruses are grouped into classes I through VI and have many variations. Some have double-stranded DNA (dsDNA), single-stranded DNA (ssDNA), single-stranded RNA (ssRNA), and double-stranded RNA (dsRNA) [9]. The viruses vary in the types of hosts they infect, the length of their genomes, size, shape, and whether or not they possess a protein shell known as capsid enclosing their genetic material. Some viruses only infect human hosts, bacteria, or animals. In general, viruses use either the lysogenic or lytic process during their replication [10].
Some viruses have only a lytic cycle, similar to a T4 bacteriophage, where the phage invades the host by binding to a specific protein and then inserts its genetic material into the host’s cell [9]. The host’s DNA is hydrolyzed, and then the phage DNA leads to the creation of the phage proteins and replication of the phage genome. The page is then assembled within the host and then produces an enzyme that creates the lysis of the host’s cell wall. When this process takes place, there can be 100 or more particles released on average during lysis [10].
The lysogenic process is similar to a lambda phage that coexists with the bacterial host [9]. The phage binds to the receptor of the host, and using a shorter tail inserts the genetic material into the host resulting in a small circle adjacent to the host DNA. A protein will then break down the host DNA. Then, the phage’s DNA is incorporated into the host DNA at a specific location, and then, both DNAs are reassembled into one [9]. The lysogenic virus has a repressor gene and can lay dormant within the host and produces a repressor protein to inhibit transcription until a later date with the host cell still able to replicate. However, environmental factors will trigger the lytic cycle of the phage DNA [9]. This process can occur in humans as well, but instead of lysogeny, it is called latency for humans, and a typical example of a virus that uses latency is Herpes Simplex Virus, where stress can trigger a Herpes outbreak [9].
Retroviruses
Retroviruses are a type of virus that utilizes RNA as its genomic material, and upon infection, they integrate into the host genome. Retroviruses are in class VI of animal viruses and exist as enveloped [10]. They enter the host’s cells through receptor-mediated endocytosis or direct diffusion. An example of the receptor-mediated process is the utilization of the receptors CD4, C-C chemokine receptor 5 (CCR5), and C-X-C chemokine receptor 4 (CXCR4) in HIV [11]. Upon entry into cells, they uncoat the capsids and integrate into the host cells’ genome using reverse transcriptase and integrase enzymes [10,12].
Retroviruses begin their lifecycle as budding from the host cell in first a non-infectious formation. As a result of various proteolytic cleavages by the enzyme protease, a structural rearrangement of the formation occurs, which allows it to achieve maturity. At this stage, it can infect new cells. The maturation processes involve changing the structure of the protein domains, correlatively this alters the conformation of the viral components that are created by the proteins [13]. The major structural component of retroviruses is the viral polyprotein, Gag, with three conserved domains for retroviruses: N-terminal matrix domain, bipartite capsid domain, and the nucleic acid-binding nucleocapsid domain (HIV GAG). To form the noninfectious immature retroviral elements, this process is driven by Gag by utilizing the nucleic acid/membrane for grounding. HIV-1, as an example, forms a hexametric lattice, which occurs in most retroviruses except for betaretroviruses and spumaviruses [13,14]. The majority of information relating to the structural aspects of retroviruses is a result of NMR, studies of protein and oligomeric domains, and studies of the latter domains and viral particles using electron microscopy [15].
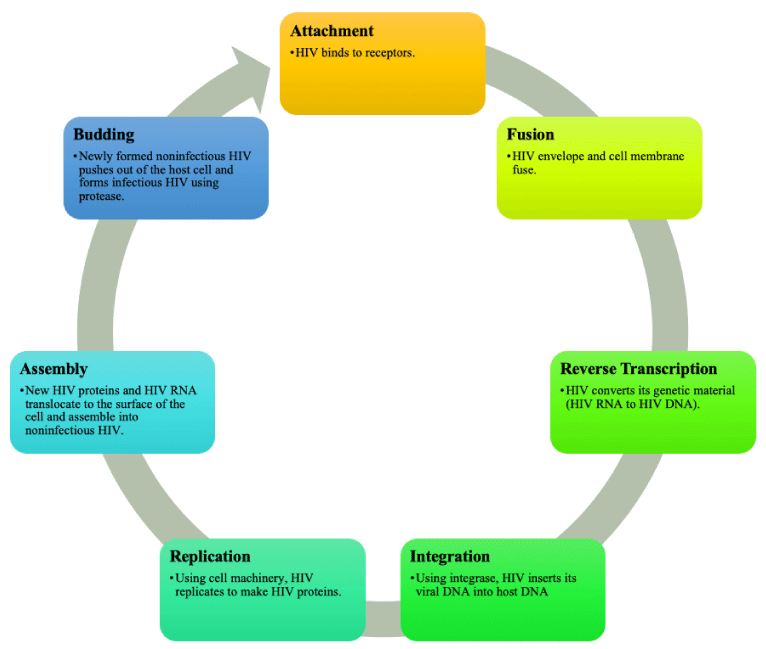
Possessing an enzyme known as reverse transcriptase, retroviruses acquire the ability to manipulate and reverse the central dogma, changing the information flow from RNA to DNA to protein in a process known as reverse transcription [16,17]. The most common organization within the genome of a retrovirus includes group-specific antigen (gag), which encodes specific structural proteins for the virus, polymerase (pol), which codes for transcriptase, protease, and integrase, and ends with envelope (env) [16,18]. Reverse transcription is essential to retroviruses as their genetic material, RNA, clashes with the DNA form of their host’s genetic material [16,18]. These retroviruses have a seven-step life cycle, beginning with binding, fusion, reverse transcription, integration, replication, assembly, and, finally, budding [19].
Attaching their surface glycoproteins to plasma membrane receptors on the host cell, the virus is fused with the cell membrane [16]. Upon infecting a host cell, reverse transcription must begin as the RNA from the retrovirus, now within the cytoplasm of the host, is required to synthesize a DNA version from the RNA molecule to integrate into the genetic material of the host cell [18]. Reverse transcriptase performs the process of producing cDNA, from an RNA template, and, once the DNA copy enters the nucleus of the host cell, another retroviral enzyme called integrase becomes involved. To incorporate the retroviral DNA, now called a provirus, into the host genome, integrase inserts the copy into the large DNA molecules of the host cell so that it is integrated into the genetic material of the host [20]. This results in a double-stranded DNA copy of the viral genome with the encoded proteins and RNA being expressed through the proviral DNA by accessing and utilizing the transcription and translation mechanisms, such as polymerases, of the host cell [16]. Now containing a new gene, the modified host cell will reproduce, and each daughter cell will possess the retroviral gene [16,18]. Translated by the host cell’s ribosomes, viral information and particles can also be released from the host cell through budding on the plasma membrane [16]. Permanently residing within the host cell, provirus infection, unlike that of bacteriophages or other viruses, is enduring and perpetual.
Zoonosis is the transmission of an infectious disease to humans from pathogens such as viruses. It is recognized that greater than 50 percent of all human infectious diseases are zoonotic, of which, the majority have emerged from the transmission of RNA viruses from animals to humans [21]. Of the many zoo tonic RNA viruses discovered, there are two orders that account for the reservoirs. Two mammalian groups, rodents (order Rodentia) and bats (order Chiroptera) are present globally and abundantly and harbor many of the zoonotic RNA viruses [21]. A majority of rodent-borne viruses are members of Hantaviridae and Arenaviridae and RNA virus sequences identified in bats include Astroviridae, Coronaviridae, Circoviridae, Adenoviridae, Filoviridae, Parvoviridae, Poxviridae, Picornaviridae, and Rhabdoviridae [21]. Zoonosis is the transmission of pathogens to humans, but humans are also capable of transmitting infectious diseases, which is known as reverse zoonosis or zooanthroponosis.
Of all of the potential zoonosis transmission methods, human-to-animal transmission is the least studied due to its rare occurrence prior to the COVID-19 pandemic [22]. Overall, for viruses, reverse zoonosis studies have mainly focused on influenza, and zooanthroponosis is yet to be reported in retroviruses [23]. Despite the limited information available on a human to animal transmission, zooanthroponosis has the potential to significantly increase the disease burden in animals with detrimental effects on wildlife as well as the potential to reinfect humans [24]. In addition to coronavirus, measles virus, influenza virus, and herpes simplex 1 virus have also been shown to possess zooanthroponic potential [24]. Human and animal interactions have been studied to better understand reverse zoonosis which has shown that domestic animals are more at risk for transmission due to their proximity to humans [24]. Despite the limited knowledge on zooanthrponosis, research on reverse zoonosis may reveal retroviruses that are able to exhibit human-to-animal transmission.
Bacterial retroviruses
There is a great diversity among retroviruses, though determining the exact age of many of them is difficult. In a host genome, the divergence between the 5’ and 3’ ends of long terminal repeats (LTRs) generated by the replication of a retrovirus has been measured in an attempt to estimate how long ago the retrovirus integrated itself into the genome [20,25]. New research indicates a possible marine origin for retroviruses that occurred about 460 to 550 million years ago [25,26]. Prokaryotes, such as bacteria and archaea, have environments that mostly hold viruses containing dsDNA genomes but have a smaller population of retroviruses. Eukaryotes such as plants and animals have environments that are much better suited for the inclusion of retroviruses, in addition to retroviruses that are either infrequent or cannot be found in prokaryotes [4,27].
Viruses can infect either prokaryotic hosts or eukaryotic hosts and those that attack bacteria are classified as bacteriophages and possess two different reproductive mechanisms. The first process is the lytic cycle which ultimately results in the death of the host cell as the phage conquers the host cell, reproduces new phages, and destroys the cell. Pages that utilize this method are classified as virulent phages. The five stages of this process include attachment, penetration, biosynthesis, maturation, and lysis. Firstly, the phage proceeds through the attachment step of this process by seeking and interacting with surface receptors on the bacteria. During penetration, the bacteriophage utilizes a tail sheath, which transports and injects the genome of the virus through the membrane of the host cell. Utilizing special, synthesized enzymes called endonucleases, the phage is then able to alter the bacterial chromosome and proceeds to replicate, transcribe, and translate essential components for the creation of new virions during the biosynthesis phase. The new phages are produced during the maturation phase and are released outside of the host cell by a process called lysis [19].
Plant-based retroviruses
In plants, under the category of transposable elements (TEs), LTR retrotransposons represent a large portion of genomic DNA sequence makeup being upwards of 17% of the whole sequence. In particular, the long terminal repeat-retrotransposons (LTR-RTs) are among the most abundant [28]. To mediate the impacts of these LTRs, molecular mechanisms exist such as DNA methylation, small mediated-RNA silencing, recombination, as well as nucleotide deletion [29]. These repeats are very effective in their incorporation into plant host genomes through the processes of transcription of the mother strand, reverse transcription, followed by the integration of that repeat into the plant’s genome.
While the proteins that are produced through the gag-pol sequence can perform both replication and transposition, other proteins have been identified that have coding capacities such as fragments of ATPase, 1,4-b-xylan endohydrolase, and 1,3-b-glucanase sequences. The sequences that are degraded or lack the ability to code for proteins, the nonautonomous elements, have been found to utilize the retrotransposons amplification processes to aid in their continued amplification and proliferation [30].
The TEs are divided into two main categories: Class I or retrotransposons and class II or DNA transposons. Class I retrotransposons utilize an RNA intermediate for transposition, while class II DNA transposons do not, a “copy and paste” versus a “cut and paste” like methodology, respectively [31]. Additionally, they are further subdivided into either LTR repeat retrotransposons or non-LTR retrotransposons. The non-LTR category contains both long interspersed repetitive elements (LINEs) as well as short interspersed repetitive elements (SINEs), with long terminal repeat retrotransposons being in higher levels in plants, while the non-LTR retrotransposons being in the highest levels, regarding the transposable element abundance [29]. In comparison to retroelements such as SINEs and LINEs, LTR retrotransposons are much more effective at spreading and integrating into plant genomes, for an unknown reason that warrants more research.
Even though the infection of a plant by a retrovirus has not been observed, because of sequencing technology, there is evidence for the integration. The LTR-RTs in plants were shown to possess an env-like gene that codes for a membrane protein in the retroviruses or plant endogenous retroviruses [32,33]. Classification of these plant retroviruses is devised according to the sequence homology and the internal of internal ORFs. The main two are the Ty1/Copia (INT-RT-RH), and Ty3/gypsy (RTRH-INT) subfamilies. Ty1/Copia has the linages of Bianca, TAR, Angela, Ale, Ivana, and Maximus, while Ty3/gypsy has the linages of Tekay, Reina, CRM, Athila, and Tat [34-36]. Current research shows that these lineages are common among both dicots and monocots, which would mean that the integration of these sequences would have occurred before the diversion of the two groups, which occurred around 140-150 million years ago [37]. Furthermore, the env-like gene, in certain families, belongs to the two gypsy and one copia lineage. A study found that the copia env-like gene was possibly brought about from inter-element recombination with the gypsy lineage, which would be significant as then they would both have a single common ancestor [36]. In plants, four main clades exist under the chromoviruses: Tekay, CRM, Galadriel, and Rina [38]. As of now, it is suggested that evolutionarily some LTR retrotransposons elements may have transferred via horizontal transmission between fungi and mosses/lyphyes, with an alternative theory being vertical transfer [38].
Endogenous retrovirus
The endogenous retroviruses (ERVs) make up to 8% of the human genome and insert their genome into the host, resulting in the expression of the retrovirus DNA within the host. Additionally, this leads to the replication of more viruses by the host. The name “endogenous retrovirus” means that the virus is inherited via the host’s DNA within the nucleus of the cell. This type of retrovirus inserts itself often within the germline cells of the host, which allows the virus to be passed onto the progeny, thus carrying on across generations [39]. There is much evidence of this process taking place throughout human evolution, where ancient retroviruses can be found within our genome today. It is likely that these retroviruses have integrated into regulatory sequences of the genome and have influenced the expression of genes within those functional coding regions [40].
The capability to sequence whole animal genomes has led to the discovery of ancient ERVs. For example, it is currently believed that human endogenous retroviruses (HERVs) may have colonized the mammalian genome between 3 million years ago or with some studies showing appearance after the hominoid and ape divergence. These insertions into the genomes have functional effects today that are alerting the expression of different genes and possible linkage to various types of cancers and autoimmune diseases, acting as superantigens within the human body [41].
The foamy viruses (FVs) are a type of retrovirus with a lasting durable history of co-speciation with mammalian hosts, specifically placental mammals within the past 100 million years. Three main mammalian endogenous FVs include Daubentonia madagascariensis (PSFVaye), Choloepus hoffmanni (SloEFV), and Chrysochloris asiatica (ChrEFV) [26]. These viruses have been associated with zoonotic transmission transmitted to humans through retroviral infection by bodily fluid contact. This was studied in Cameroon, Africa with local villagers with retroviral sequences being discovered in their genomes, thus an example of modern-day integration [42]. Similarly, in Asia, a similar transmission pattern exists between humans and macaques species at specific temple sites, showing transmission of these FVs existing in modern times between primates and humans [43].
At the molecular level, the ERVs play a role in human development through the regulation of neuronal progenitor cells (NPCs), based upon the Tripartite motif-containing protein 28 (TRIM28), and serving to regulate the expression of an ERV through a repression mechanism. This consequently regulates the expression of other genes [44]. The replication of Moloney Murine Leukemia Virus expression during embryogenesis is known to be silenced due to the TRIM28 protein to prevent and protect the germ line from mutagenesis. There is an upregulation in the expression of many different ERVs once TRIM28 was deleted, with a mild upregulation in retroelements like LINE-1 but more significant upregulation of some ERVs such as the Mus musculus ERV and intracisternal A-particles class 1 (IAP1) as well as other IAPs, though not all [45].
The ERV elements can be controlled epigenetically through histone modifications and methylations [46]. It was found in the HML-2 (a subgroup of HERV-K) that expression was high in tissues of the placenta and testicles due to hypomethylation in LTRs and androgens stimulating expression [47]. For the HERVs to be functional, the LTR must not be disrupted by nucleotide substitution or deletion within the open reading frame, which can lead to the control of expression being in another promoter region. As such, when epigenetics is affected by stimuli, it can also lead to ERV proteins being transcribed. Epigenetics can be disrupted by factors such as inflammation, Herpesviridae, HLTV-1 Tax transactivator, and HIV Tat protein, all of which can lead to the upregulation of transcription of ERVs [46].
Human retroviruses are abundant with their LTRs which can be found throughout the genome and can have a significant impact on the expression of neighboring genes. In fact, a study found that p53, a tumor suppressor protein, directly binds to the binding sites in LTRs specifically of the HML-2 sup-group of the HERV-K family [48]. In addition, HERVs have a strong association with various cancers. For example, HERV-K is strongly associated with neurological, prostate, mammary, lung, and skin cancers including Kaposi’s sarcoma. Pituitary adenoma, pancreatic, urological, gynecological, and hematological cancers are also associated with HERVs [49]. Additionally, in recent years, retroviral oncogenes have also been identified [50]. The HERVs silenced by mutations and recombination have also been reported to have higher levels of expression in various cancers due to dysregulated gene expression [51].
Human retroviruses
Human retroviruses, in addition to HERVs, belong to different genera and classes and have a wide range of implications on their host. Table 1 shows the select genera and types of retroviruses that infect humans. The primer-binding site (PBS) sequence has traditionally been used for HERV nomenclature, where the group names are identified using a letter that characterizes the human tRNA type and binds to the viral PBS sequences. If the PBS sequence is not available, HERVs are named according to unconventional methods such as the name of the neighboring gene, a clone number, or an amino acid motif [52].
Human T-cell leukemia virus
The human T-cell leukemia virus (HTLV) is a complex retrovirus that belongs to the Deltaretrovirus family [53,54]. This virus is considered a zoonotic virus with simian T-lymphotropic viruses (STLVs) [55]. The HTLV route of transmission is based on the zoonosis of STLVs from monkeys to humans around 30,000 to 40,000 years ago, which lead to the evolution of STLV to HTLV [52]. It is formed from a single positive strand of RNA, and it primarily infects T-lymphocytes [56]. In general, it is transmitted horizontally from humans to other humans and is associated with rare diseases, such as neuroinflammatory disease HTLV-1–associated myelopathy/tropical spastic paraparesis (HAM/TSP) and adult T-cell leukemia/lymphoma [57,58].
HTLV-1 is one of the most common retroviruses in the HTLV family [56]. HTLV-2 is another subtype of HTLV and, along with HTLV-1, is one of the most researched HTLV subtypes. HTLV-2 is found in hairy T-cell leukemia and is currently not associated with lymphoproliferative disease [55]. HTLV-1 is mainly identified in CD4+ T-lymphocytes and HTLV-2 is common in CD8+ T-lymphocytes.
Human immunodeficiency virus and acquired immunodeficiency syndrome
Human immunodeficiency virus (HIV) and acquired immunodeficiency syndrome (AIDS) are retroviruses targeting human white blood cells, specifically CD4 cells and thereby weakening the host immune system [59]. The first case of HIV was reported in 1981. Globally, 38.4 million individuals were living with HIV in 2021 [59]. An estimated 1,189,700 individuals in the United States had HIV by the end of 2019 with 30,635 new diagnoses in 2020 [60]. Most HIV infections are caused by HIV type 1 and a smaller percentage of infections are caused by HIV type 2 [2]. The life cycle of HIV is shown in Figure 2.
Though HIV targets the immune system, the effects of HIV are profound with significant effects on the central nervous system [62]. Symptoms are often addressed through various therapeutics such as cannabinoids to address appetite and anti-emesis [63]. Antiretroviral therapy (ART) and personalized medicine have reduced mortality and morbidity of HIV and AIDS, but more efficient alternatives for therapy such as clustered regularly interspaced short palindromic repeats (CRISPR) and vaccines are being explored [64-67].
Applications and personalized medicine
Personalized medicine allows for customized treatment plans, earlier interventions, and efficient drug therapies for each patient. Utilizing the genome of an individual, doctors can prevent, diagnose, and treat diseases [68]. Pharmacogenomics uses the genome of the patient to determine the best type of drug treatments and understand the signaling pathways involved in the specific subtypes to determine the efficacy of drug-based treatments. In terms of retroviruses, it is possible to use gene therapy as a method of treatment using personalized medicine based on the sequences. It can be used with less risk and greater safety by targeting the patient’s cells [69]. Gene therapy, which is the treatment of diseases by targeting specific genes in a patient’s cells, can be used to treat patients with greater safety and reduce the problems associated with drug treatments [69].
An HIV remission case study suggests that ART is used to treat HIV in an infected individual though it requires lifelong treatment [70]. The first known HIV-1 remission case was in 2009 with a patient known as the “Berlin Patient” and the second patient was known as the “London Patient” [70]. Hematopoietic stem cell transplantation (HSCT) allows patients to restore their immune systems. However, after the transplant, patients stop taking their antiretrovirals allowing for the potential of viral infection. However, the stem cell transplant to treat the patient’s cancer resulted in HIV going into remission. The HIV became undetectable even though the patient was no longer on antiretroviral therapy for at least 16 months. Currently, the London Patient may be in remission; however, they currently have a low level of Plasma HIV, RNA, or DNA that is insignificant in the London Patient’s and Berlin Patients’ blood. The researchers believe the London Patient could have a relapse; however, they will know more in 6 months without the patient on antiretroviral treatments. The factors that caused long-term remission of HIV-1 are not fully understood and are further convoluted due to the differences in the two remission cases [70]. Both patients did experience other complications at the hand of long-term remission. Currently, there is no precise HIV treatment that allows for long-term remission. However, researchers are looking further into the C-C chemokine receptor type 5 (CCR5) gene, which is known to be involved in allowing HIV to enter host cells, and hopefully, soon, there will be double-blind studies for treatment for HIV that allow long-term remission. Though ART has lowered the mortality of HIV/AIDS, there are pitfalls to ART including detrimental effects on the nervous system such as penetrating the blood-brain barrier and nervous system toxicity [71]. Additionally, ART is not able to protect against latent infections of HIV which may go undetected [71].
CRISPR/Cas9
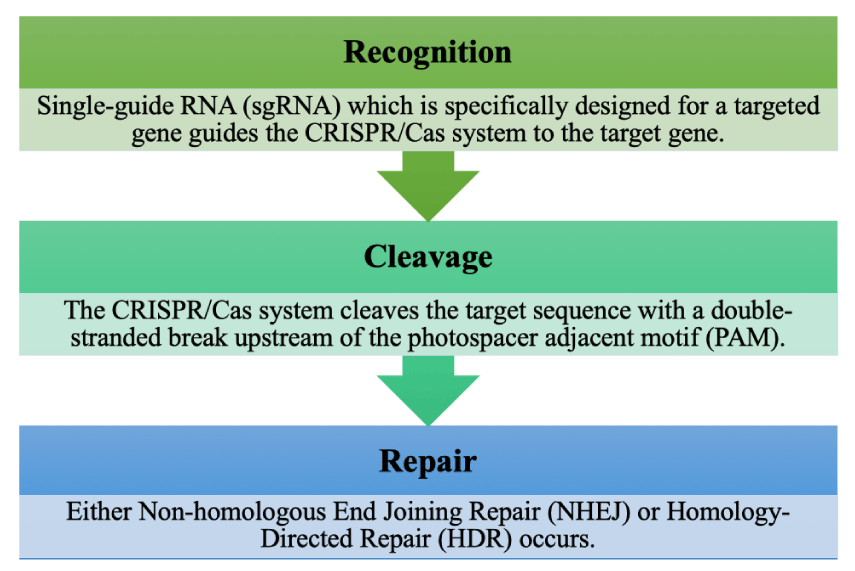
The use of CRISPR is a new genome-editing technology that has allowed for increased speed, efficiency, accuracy, and reduced cost compared to previous technologies [72]. Prior genome editing techniques expensive, difficult to make, and error-prone, such as zinc-finger nucleases (ZFNs) and Transcription Activator-Like Effector Nucleases (TALENs). CRISPR is used as an active immune system by bacteria to defend against bacteriophages and works with CRISPR-associated (Cas) genes like Cas9 [73]. This Cas9 enzyme is what gives bacteria this resistance by cutting and disabling the DNA of the bacteriophage as shown in Figure 3 [74]. If there is no matching spacer for the bacteriophage DNA, a different Class 1 Cas enzyme is created that cuts the bacteriophage DNA and inserts it into a spacer between the palindromic repeating sequences, which prevents future infection from the bacteriophage [73,75].
Though CRISPR has been utilized in plants [76-78], more recently, CRISPR has been used for human ailments such as sickle cell anemia and cancer. Sickle cell anemia is a genetic disease where an individual has sickle-shaped erythrocytes. Sickle cells are caused by a change in a single amino acid, where glutamic acid is replaced with valine [79]. The bone marrow cells were removed and CRISPR was used to edit fetal hemoglobin to activate it [80]. Once the fetal hemoglobin is active, it should compensate for and over time, replace the adult sickle cells. More than 2 billion cells were edited and returned to the patient’s body. Additionally, the initial blood work was reviewed, which showed the edited cells are working, and the fetal hemoglobin is beginning to be produced. Almost half of the patient’s hemoglobin is fetal and appears to be increasing.
The University of Pennsylvania has had a clinical trial approved for to use of CRISPR gene editing for cancer patients in the first safety trial [80,81]. The test was meant to determine if CRISPR gene editing worked and if it was safe to use. Of the three cancer patients involved, two had multiple myeloma, and one with sarcoma received genetically edited cells. All patients were screened for the antigen HLA-A201, which allows for the CRISPR gene to become active. The patients had 100 million T cells that were removed from the patient blood and then edited with CRISPR. Patients were also treated with chemotherapy and then received a single infusion of the edited genes. CRISPR was used to remove three genes, first targeting the T-cell receptor α chain (TRAC) gene and second targeting the T-cell receptor β chain (TRBC) gene [81]. In either case, the T-cell receptors would have been removed, so they bind appropriately to the cancer cells. The results showed high efficiency for TRAC and low efficiency for TRBC [81]. The third gene to be removed was programmed cell death protein 1 (PDCD-1) which encodes for an immune checkpoint protein, which at times can block T cells from their function [81]. The function of the receptor is to have the T-cells target the antigen NY-ESO-1, which would allow the body to attack the specified cancer cells more effectively and aggressively. The patients were treated in January, April, and August, and the first patient was followed for six months and showed no significant side effects [81]. All three patients’ blood samples showed that the T-cells expanded and survived. The T-cells expand and bind to their tumor targets [81]. The doctors explain the results show promise, but patients will need to have constant and longer follow-ups to check for side effects, and these results are still early to be conclusive [81].
CRISPR has also been utilized with retroviruses notably for HIV and HTLV-1. The utilization of CRISPR for HIV treatment has been studied extensively in recent years with multiple different therapeutic strategies [65,66]. CRISPR/Cas9 system does have the ability to cleave the HIV genome rendering the virus nonfunctional with multiple studies indicating successful cleavage of proviral HIV DNA [64]. Additionally, CRISPR can be used to cleave either the LTR domain with one single guide RNA (sgRNA), specifically the LTR domain, or cleave two different areas of the genome using two different sgRNAs to introduce mutations to inactivate the HIV provirus [64,82]. CRISPR can also be used as a defense mechanism against HIV by targeting the newly formed double-stranded DNA from reverse transcription [64,83,84]. However, there is a potential for HIV to avoid detection by CRISPR. Alternatives such as combinatorial use of CRISPR to result in hypermutation or combining CRISPR with traditional therapeutics such as ART have been explored [64,84-86]. A recent case study successfully used CRISPR-edited hematopoietic stem and progenitor cells (HSPCs) that were transplanted into a patient with HIV and acute lymphoblastic leukemia [87]. Additionally, a study found that CRISPR/Cas13a system was able to inhibit HIV-1 viral infection [88].
CRISPR utilization for HTLV-1 is a relatively new concept. The foundation of the potential usage of CRISPR for therapeutic advantages in HTLV-1 comes from the success of the use of CRISPR in sickle cell anemia and cancer [89]. Additionally, support from other gene editing technologies such as ZFNs showed that gene editing results in the disruption of proliferation that is regulated by HTLV-1 [89-92]. This information opens a potential avenue for CRISPR-mediated therapy for HTLV-1 likely in the form of CRISPR delivery in vivo [89,93].
Conclusion
Through the study and discovery of retroviruses, the Central Dogma has been modified and amended. The knowledge and the diversity of retroviruses have demonstrated vast implications across kingdoms and all evolutionary ranks. The study of retroviruses has also had a considerable impact on the diverse field of biology. The repercussions of retroviruses on human health are exemplified by cases of HIV, HTLV, and related morbidities. The magnitude of the impact of retroviruses experienced across the kingdoms highlights the importance of developing a better understanding of retroviruses, their role in genome diversity, and their effects on the host organism. With the advent of new technology such as CRISPR, the latest therapeutic avenues are being explored for retroviral infections and associated morbidities such as cancer.
The authors would like to acknowledge the School of Science, Engineering, and Technology at Penn State Harrisburg for their support.
- Crick F. Central dogma of molecular biology. Nature. 1970 Aug 8;227(5258):561-3. doi: 10.1038/227561a0. PMID: 4913914.
- Meissner ME, Talledge N, Mansky LM. Molecular Biology and Diversification of Human Retroviruses. Front Virol. 2022;2:872599. doi: 10.3389/fviro.2022.872599. Epub 2022 Jun 2. PMID: 35783361; PMCID: PMC9242851.
- Sanjuan R, Illingworth CJR, Geoghegan JL, Iranzo J, Zwart MP, Ciota AT, Moratorio G, Gago-Zachert S, Duffy S, Vijaykrishna D. Five Challenges in the Field of Viral Diversity and Evolution. Frontiers in Virology, 4. 2021.
- Dolja VV, Koonin EV. Common origins and host-dependent diversity of plant and animal viromes. Curr Opin Virol. 2011 Nov;1(5):322-31. doi: 10.1016/j.coviro.2011.09.007. PMID: 22408703; PMCID: PMC3293486.
- Martin W, Koonin EV. Introns and the origin of nucleus-cytosol compartmentalization. Nature. 2006 Mar 2;440(7080):41-5. doi: 10.1038/nature04531. PMID: 16511485.
- Koonin EV, Senkevich TG, Dolja VV. The ancient Virus World and evolution of cells. Biol Direct. 2006 Sep 19;1:29. doi: 10.1186/1745-6150-1-29. PMID: 16984643; PMCID: PMC1594570.
- Koonin EV. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J Gen Virol. 1991 Sep;72 ( Pt 9):2197-206. doi: 10.1099/0022-1317-72-9-2197. PMID: 1895057.
- Eickbush TH, Jamburuthugoda VK. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 2008 Jun;134(1-2):221-34. doi: 10.1016/j.virusres.2007.12.010. Epub 2008 Feb 7. PMID: 18261821; PMCID: PMC2695964.
- Dimmock NJ, Easton AJ, Leppard KN. Introduction to Modern Virology. Wiley Blackwell. 7th Edition. Chichester, West Sussex. 2016.
- Young R. Bacteriophage lysis: mechanism and regulation. Microbiol Rev. 1992 Sep;56(3):430-81. doi: 10.1128/mr.56.3.430-481.1992. PMID: 1406491; PMCID: PMC372879.
- Baleux F, Loureiro-Morais L, Hersant Y, Clayette P, Arenzana-Seisdedos F, Bonnaffé D, Lortat-Jacob H. A synthetic CD4-heparan sulfate glycoconjugate inhibits CCR5 and CXCR4 HIV-1 attachment and entry. Nat Chem Biol. 2009 Oct;5(10):743-8. doi: 10.1038/nchembio.207. Epub 2009 Sep 6. PMID: 19734912.
- Beemon KL. Retroviral RNA Processing. Viruses. 2022 May 23;14(5):1113. doi: 10.3390/v14051113. PMID: 35632854; PMCID: PMC9143442.
- Mattei S, Schur FK, Briggs JA. Retrovirus maturation-an extraordinary structural transformation. Curr Opin Virol. 2016 Jun;18:27-35. doi: 10.1016/j.coviro.2016.02.008. Epub 2016 Mar 22. PMID: 27010119.
- Waheed AA, Freed EO. The Role of Lipids in Retrovirus Replication. Viruses. 2010 May 1;2(5):1146-1180. doi: 10.3390/v2051146. PMID: 20740061; PMCID: PMC2927015.
- Zhang W, Cao S, Martin JL, Mueller JD, Mansky LM. Morphology and ultrastructure of retrovirus particles. AIMS Biophys. 2015;2(3):343-369. doi: 10.3934/biophy.2015.3.343. Epub 2015 Aug 18. PMID: 26448965; PMCID: PMC4593330.
- Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; Overview of Reverse Transcription. 1997. https://www.ncbi.nlm.nih.gov/books/NBK19424/
- Sáez-Cirión A, Manel N. Immune Responses to Retroviruses. Annu Rev Immunol. 2018 Apr 26;36:193-220. doi: 10.1146/annurev-immunol-051116-052155. Epub 2018 Jan 12. PMID: 29328787.
- Grandi N, Tramontano E. Human Endogenous Retroviruses Are Ancient Acquired Elements Still Shaping Innate Immune Responses. Front Immunol. 2018 Sep 10;9:2039. doi: 10.3389/fimmu.2018.02039. PMID: 30250470; PMCID: PMC6139349..
- Reece JB, Murry LA, Curry ML, Wasserman SA, Minorsky PV, Jackson RB. Viruses. In Campbell Biology. 10th Edition. Boston: Pearson. 2014.
- Gifford RJ, Katzourakis A, Tristem M, Pybus OG, Winters M, Shafer RW. A transitional endogenous lentivirus from the genome of a basal primate and implications for lentivirus evolution. Proc Natl Acad Sci U S A. 2008 Dec 23;105(51):20362-7. doi: 10.1073/pnas.0807873105. Epub 2008 Dec 15. PMID: 19075221; PMCID: PMC2603253.
- Williams EP, Spruill-Harrell BM, Taylor MK, Lee J, Nywening AV, Yang Z, Nichols JH, Camp JV, Owen RD, Jonsson CB. Common Themes in Zoonotic Spillover and Disease Emergence: Lessons Learned from Bat- and Rodent-Borne RNA Viruses. Viruses. 2021 Jul 31;13(8):1509. doi: 10.3390/v13081509. PMID: 34452374; PMCID: PMC8402684.
- Jia P, Dai S, Wu T, Yang S. New Approaches to Anticipate the Risk of Reverse Zoonosis. Trends Ecol Evol. 2021 Jul;36(7):580-590. doi: 10.1016/j.tree.2021.03.012. Epub 2021 May 6. PMID: 33966919; PMCID: PMC8100872.
- Messenger AM, Barnes AN, Gray GC. Reverse zoonotic disease transmission (zooanthroponosis): a systematic review of seldom-documented human biological threats to animals. PLoS One. 2014 Feb 28;9(2):e89055. doi: 10.1371/journal.pone.0089055. PMID: 24586500; PMCID: PMC3938448.
- Epstein JH, Price JT. The significant but understudied impact of pathogen transmission from humans to animals. Mt Sinai J Med. 2009 Oct;76(5):448-55. doi: 10.1002/msj.20140. PMID: 19787650; PMCID: PMC7168516.
- Hayward A. Origin of the retroviruses: when, where, and how? Curr Opin Virol. 2017 Aug;25:23-27. doi: 10.1016/j.coviro.2017.06.006. Epub 2017 Jun 30. PMID: 28672160; PMCID: PMC5962544.
- Aiewsakun P, Katzourakis A. Marine origin of retroviruses in the early Palaeozoic Era. Nat Commun. 2017 Jan 10;8:13954. doi: 10.1038/ncomms13954. PMID: 28071651; PMCID: PMC5512871.
- Pina M, Bize A, Forterre P, Prangishvili D. The archeoviruses. FEMS Microbiol Rev. 2011 Nov;35(6):1035-54. doi: 10.1111/j.1574-6976.2011.00280.x. Epub 2011 Jun 6. PMID: 21569059.
- McCarthy EM, Liu J, Lizhi G, McDonald JF. Long terminal repeat retrotransposons of Oryza sativa. Genome Biol. 2002 Sep 13;3(10):RESEARCH0053. doi: 10.1186/gb-2002-3-10-research0053. Epub 2002 Sep 13. PMID: 12372141; PMCID: PMC134482.
- Zhao M, Ma J. Co-evolution of plant LTR-retrotransposons and their host genomes. Protein Cell. 2013 Jul;4(7):493-501. doi: 10.1007/s13238-013-3037-6. Epub 2013 Jun 23. PMID: 23794032; PMCID: PMC4875514.
- Kalendar R, Vicient CM, Peleg O, Anamthawat-Jonsson K, Bolshoy A, Schulman AH. Large retrotransposon derivatives: abundant, conserved but nonautonomous retroelements of barley and related genomes. Genetics. 2004 Mar;166(3):1437-50. doi: 10.1534/genetics.166.3.1437. PMID: 15082561; PMCID: PMC1470764.
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, Paux E, SanMiguel P, Schulman AH. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007 Dec;8(12):973-82. doi: 10.1038/nrg2165. PMID: 17984973.
- Wright DA, Voytas DF. Athila4 of Arabidopsis and Calypso of soybean define a lineage of endogenous plant retroviruses. Genome Res. 2002 Jan;12(1):122-31. doi: 10.1101/gr.196001. PMID: 11779837; PMCID: PMC155253.
- Laten HM, Majumdar A, Gaucher EA. SIRE-1, a copia/Ty1-like retroelement from soybean, encodes a retroviral envelope-like protein. Proc Natl Acad Sci U S A. 1998 Jun 9;95(12):6897-902. doi: 10.1073/pnas.95.12.6897. PMID: 9618510; PMCID: PMC22677.
- Wicker T, Keller B. Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families. Genome Res. 2007 Jul;17(7):1072-81. doi: 10.1101/gr.6214107. Epub 2007 Jun 7. PMID: 17556529; PMCID: PMC1899118.
- Wang W, Zheng H, Fan C, Li J, Shi J, Cai Z, Zhang G, Liu D, Zhang J, Vang S, Lu Z, Wong GK, Long M, Wang J. High rate of chimeric gene origination by retroposition in plant genomes. Plant Cell. 2006 Aug;18(8):1791-802. doi: 10.1105/tpc.106.041905. Epub 2006 Jul 7. PMID: 16829590; PMCID: PMC1533979.
- Du J, Tian Z, Hans CS, Laten HM, Cannon SB, Jackson SA, Shoemaker RC, Ma J. Evolutionary conservation, diversity and specificity of LTR-retrotransposons in flowering plants: insights from genome-wide analysis and multi-specific comparison. Plant J. 2010 Aug;63(4):584-98. doi: 10.1111/j.1365-313X.2010.04263.x. PMID: 20525006.
- Chaw SM, Chang CC, Chen HL, Li WH. Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J Mol Evol. 2004 Apr;58(4):424-41. doi: 10.1007/s00239-003-2564-9. PMID: 15114421.
- Novikova O, Smyshlyaev G, Blinov A. Evolutionary genomics revealed interkingdom distribution of Tcn1-like chromodomain-containing Gypsy LTR retrotransposons among fungi and plants. BMC Genomics. 2010 Apr 8;11:231. doi: 10.1186/1471-2164-11-231. PMID: 20377908; PMCID: PMC2864245.
- Bock M, Stoye JP. Endogenous retroviruses and the human germline. Curr Opin Genet Dev. 2000 Dec;10(6):651-5. doi: 10.1016/s0959-437x(00)00138-6. PMID: 11088016.
- Khodosevich K, Lebedev Y, Sverdlov E. Endogenous retroviruses and human evolution. Comp Funct Genomics. 2002;3(6):494-8. doi: 10.1002/cfg.216. PMID: 18629260; PMCID: PMC2448423.
- Balada E, Ordi-Ros J, Vilardell-Tarrés M. Molecular mechanisms mediated by human endogenous retroviruses (HERVs) in autoimmunity. Rev Med Virol. 2009 Sep;19(5):273-86. doi: 10.1002/rmv.622. PMID: 19714703.
- Wolfe ND, Switzer WM, Carr JK, Bhullar VB, Shanmugam V, Tamoufe U, Prosser AT, Torimiro JN, Wright A, Mpoudi-Ngole E, McCutchan FE, Birx DL, Folks TM, Burke DS, Heneine W. Naturally acquired simian retrovirus infections in central African hunters. Lancet. 2004 Mar 20;363(9413):932-7. doi: 10.1016/S0140-6736(04)15787-5. PMID: 15043960.
- Engel G, Hungerford LL, Jones-Engel L, Travis D, Eberle R, Fuentes A, Grant R, Kyes R, Schillaci M. Risk assessment: A model for predicting cross-species transmission of simian foamy virus from macaques (M. fascicularis) to humans at a monkey temple in Bali, Indonesia. Am J Primatol. 2006 Sep;68(9):934-48. doi: 10.1002/ajp.20299. PMID: 16900504.
- Brattås PL, Jönsson ME, Fasching L, Nelander Wahlestedt J, Shahsavani M, Falk R, Falk A, Jern P, Parmar M, Jakobsson J. TRIM28 Controls a Gene Regulatory Network Based on Endogenous Retroviruses in Human Neural Progenitor Cells. Cell Rep. 2017 Jan 3;18(1):1-11. doi: 10.1016/j.celrep.2016.12.010. PMID: 28052240.
- Montesion M, Williams ZH, Subramanian RP, Kuperwasser C, Coffin JM. Promoter expression of HERV-K (HML-2) provirus-derived sequences is related to LTR sequence variation and polymorphic transcription factor binding sites. Retrovirology. 2018 Aug 20;15(1):57. doi: 10.1186/s12977-018-0441-2. PMID: 30126415; PMCID: PMC6102855.
- Küry P, Nath A, Créange A, Dolei A, Marche P, Gold J, Giovannoni G, Hartung HP, Perron H. Human Endogenous Retroviruses in Neurological Diseases. Trends Mol Med. 2018 Apr;24(4):379-394. doi: 10.1016/j.molmed.2018.02.007. Epub 2018 Mar 15. PMID: 29551251; PMCID: PMC7185488.
- Garcia-Montojo M, Doucet-O'Hare T, Henderson L, Nath A. Human endogenous retrovirus-K (HML-2): a comprehensive review. Crit Rev Microbiol. 2018 Nov;44(6):715-738. doi: 10.1080/1040841X.2018.1501345. Epub 2018 Oct 14. PMID: 30318978; PMCID: PMC6342650.
- Liu M, Jia L, Li H, Liu Y, Han J, Wang X, Li T, Li J, Zhang B, Zhai X, Yu C, Li L. p53 Binding Sites in Long Terminal Repeat 5Hs (LTR5Hs) of Human Endogenous Retrovirus K Family (HML-2 Subgroup) Play Important Roles in the Regulation of LTR5Hs Transcriptional Activity. Microbiol Spectr. 2022 Aug 31;10(4):e0048522. doi: 10.1128/spectrum.00485-22. Epub 2022 Jul 18. PMID: 35867400; PMCID: PMC9430305.
- Müller MD, Holst PJ, Nielsen KN. A Systematic Review of Expression and Immunogenicity of Human Endogenous Retroviral Proteins in Cancer and Discussion of Therapeutic Approaches. Int J Mol Sci. 2022 Jan 25;23(3):1330. doi: 10.3390/ijms23031330. PMID: 35163254; PMCID: PMC8836156.
- Lipsick J. A History of Cancer Research: Retroviral Oncogenes. Cold Spring Harb Perspect Med. 2022 May 17;12(4):a035865. doi: 10.1101/cshperspect.a035865. PMID: 35581009; PMCID: PMC9121892.
- Sahu S, Singh B, Kumar Rai A. Human endogenous retrovirus regulates the initiation and progression of cancers (Review). Mol Clin Oncol. 2022 Aug 3;17(4):143. doi: 10.3892/mco.2022.2576. PMID: 36157315; PMCID: PMC9468830.
- Luganini A, Gribaudo G. Retroviruses of the Human Virobiota: The Recycling of Viral Genes and the Resulting Advantages for Human Hosts During Evolution. Front Microbiol. 2020 May 28;11:1140. doi: 10.3389/fmicb.2020.01140. PMID: 32547531; PMCID: PMC7270195.
- Nelson PN, Carnegie PR, Martin J, Davari Ejtehadi H, Hooley P, Roden D, Rowland-Jones S, Warren P, Astley J, Murray PG. Demystified. Human endogenous retroviruses. Mol Pathol. 2003 Feb;56(1):11-8. doi: 10.1136/mp.56.1.11. PMID: 12560456; PMCID: PMC1187282.
- Rosenberg N. Overview of retrovirology. In Retroviruses and insights into cancer. Springer, New York, NY. 2010; 1-30.
- Martinez MP, Al-Saleem J, Green PL. Comparative virology of HTLV-1 and HTLV-2. Retrovirology. 2019 Aug 7;16(1):21. doi: 10.1186/s12977-019-0483-0. PMID: 31391116; PMCID: PMC6686503.
- Zhang LL, Wei JY, Wang L, Huang SL, Chen JL. Human T-cell lymphotropic virus type 1 and its oncogenesis. Acta Pharmacol Sin. 2017 Aug;38(8):1093-1103. doi: 10.1038/aps.2017.17. Epub 2017 Apr 10. PMID: 28392570; PMCID: PMC5547553.
- Cloyd MW. Human Retroviruses. In S. Baron (Ed.), Medical Microbiology (4th ed.). University of Texas Medical Branch at Galveston. 1996
- Araya N, Sato T, Ando H, Tomaru U, Yoshida M, Coler-Reilly A, Yagishita N, Yamauchi J, Hasegawa A, Kannagi M, Hasegawa Y, Takahashi K, Kunitomo Y, Tanaka Y, Nakajima T, Nishioka K, Utsunomiya A, Jacobson S, Yamano Y. HTLV-1 induces a Th1-like state in CD4+CCR4+ T cells. J Clin Invest. 2014 Aug;124(8):3431-42. doi: 10.1172/JCI75250. Epub 2014 Jun 24. PMID: 24960164; PMCID: PMC4109535.
- World Health Organization. HIV/AIDS. World Health Organization. https://www.who.int/health-topics/hiv-aids#tab=tab_1 2022.
- Centers for Disease Control and Prevention. HIV Basics: Basic Statistics. Centers for Disease Control and Prevention. https://www.cdc.gov/hiv/basics/statistics.html 2022.
- National Institute of Health. HIV Overview: The HIV Life Cycle. National Institute of Health. https://hivinfo.nih.gov/understanding-hiv/fact-sheets/hiv-life-cycle#:~:text=The%20seven%20stages%20of%20the%20HIV%20life%20cycle%20are%3A%201,imagine%20what%20HIV%20looks%20like.
- Rojas-Celis V, Valiente-Echeverría F, Soto-Rifo R, Toro-Ascuy D. New Challenges of HIV-1 Infection: How HIV-1 Attacks and Resides in the Central Nervous System. Cells. 2019 Oct 13;8(10):1245. doi: 10.3390/cells8101245. PMID: 31614895; PMCID: PMC6829584.
- Breijyeh Z, Jubeh B, Bufo SA, Karaman R, Scrano L. Cannabis: A Toxin-Producing Plant with Potential Therapeutic Uses. Toxins (Basel). 2021 Feb 5;13(2):117. doi: 10.3390/toxins13020117. PMID: 33562446; PMCID: PMC7915118.
- Wang G, Zhao N, Berkhout B, Das AT. CRISPR-Cas based antiviral strategies against HIV-1. Virus Res. 2018 Jan 15;244:321-332. doi: 10.1016/j.virusres.2017.07.020. Epub 2017 Jul 29. PMID: 28760348.
- Panfil AR, Green PL, Yoder KE. CRISPR Genome Editing Applied to the Pathogenic Retrovirus HTLV-1. Front Cell Infect Microbiol. 2020 Dec 23;10:580371. doi: 10.3389/fcimb.2020.580371. PMID: 33425776; PMCID: PMC7785941.
- Darcis G, Das AT, Berkhout B. Tackling HIV Persistence: Pharmacological versus CRISPR-Based Shock Strategies. Viruses. 2018 Mar 29;10(4):157. doi: 10.3390/v10040157. PMID: 29596334; PMCID: PMC5923451.
- Burton DR. Advancing an HIV vaccine; advancing vaccinology. Nat Rev Immunol. 2019 Feb;19(2):77-78. doi: 10.1038/s41577-018-0103-6. PMID: 30560910; PMCID: PMC6425752.
- Cecchin E, Stocco G. Pharmacogenomics and Personalized Medicine. Genes (Basel). 2020 Jun 22;11(6):679. doi: 10.3390/genes11060679. PMID: 32580376; PMCID: PMC7348959.
- Tosolini AP, Smith GM. Editorial: Gene Therapy for the Central and Peripheral Nervous System. Front Mol Neurosci. 2018 Feb 22;11:54. doi: 10.3389/fnmol.2018.00054. PMID: 29527154; PMCID: PMC5829609.
- Gupta RK, Abdul-Jawad S, McCoy LE, Mok HP, Peppa D, Salgado M, Martinez-Picado J, Nijhuis M, Wensing AMJ, Lee H, Grant P, Nastouli E, Lambert J, Pace M, Salasc F, Monit C, Innes AJ, Muir L, Waters L, Frater J, Lever AML, Edwards SG, Gabriel IH, Olavarria E. HIV-1 remission following CCR5Δ32/Δ32 haematopoietic stem-cell transplantation. Nature. 2019 Apr;568(7751):244-248. doi: 10.1038/s41586-019-1027-4. Epub 2019 Mar 5. PMID: 30836379; PMCID: PMC7275870.
- Rudd H, Toborek M. Pitfalls of Antiretroviral Therapy: Current Status and Long-Term CNS Toxicity. Biomolecules. 2022 Jun 26;12(7):894. doi: 10.3390/biom12070894. PMID: 35883450; PMCID: PMC9312798.
- Varshney GK, Pei W, LaFave MC, Idol J, Xu L, Gallardo V, Carrington B, Bishop K, Jones M, Li M, Harper U, Huang SC, Prakash A, Chen W, Sood R, Ledin J, Burgess SM. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res. 2015 Jul;25(7):1030-42. doi: 10.1101/gr.186379.114. Epub 2015 Jun 5. PMID: 26048245; PMCID: PMC4484386.
- Lander ES. The Heroes of CRISPR. Cell. 2016 Jan 14;164(1-2):18-28. doi: 10.1016/j.cell.2015.12.041. PMID: 26771483.
- Asmamaw M, Zawdie B. Mechanism and Applications of CRISPR/Cas-9-Mediated Genome Editing. Biologics. 2021 Aug 21;15:353-361. doi: 10.2147/BTT.S326422. PMID: 34456559; PMCID: PMC8388126.
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008 Aug 15;321(5891):960-4. doi: 10.1126/science.1159689. PMID: 18703739; PMCID: PMC5898235.
- Deguchi M, Kane S, Potlakayala S, George H, Proano R, Sheri V, Curtis WR, Rudrabhatla S. Metabolic Engineering Strategies of Industrial Hemp (Cannabis sativa L.): A Brief Review of the Advances and Challenges. Front Plant Sci. 2020 Dec 8;11:580621. doi: 10.3389/fpls.2020.580621. PMID: 33363552; PMCID: PMC7752810.
- Mistry M, Patel A, Patel S, Potlakayala S, Rudrabhatla S. An Overview of CRISPR/Cas9 Genome Editing System on Nutraceuticals. Current Trends on Biotechnology & Microbiology. 2022; 2(4).
- Deguchi M, Dhir S, Potlakayala S, Dhir S, Curtis WR, Rudrabhatla S. In planta Female Flower Agroinfiltration Alters the Cannabinoid Composition in Industrial Hemp (Cannabis sativa L.). Front Plant Sci. 2022 Jul 21;13:921970. doi: 10.3389/fpls.2022.921970. PMID: 35941940; PMCID: PMC9356322.
- Piel FB, Patil AP, Howes RE, Nyangiri OA, Gething PW, Dewi M, Temperley WH, Williams TN, Weatherall DJ, Hay SI. Global epidemiology of sickle haemoglobin in neonates: a contemporary geostatistical model-based map and population estimates. Lancet. 2013 Jan 12;381(9861):142-51. doi: 10.1016/S0140-6736(12)61229-X. Epub 2012 Oct 25. PMID: 23103089; PMCID: PMC3547249.
- Stein R. In A 1st, Doctors In U.S. Use Tool To Treat Patient With Genetic Disorder. 2019. https://www.npr.org/sections/health-shots/2019/07/29/744826505/sickle-cell-patient-reveals-why-she-is-volunteering-for-landmark-gene-editing-st
- Stadtmauer EA, Fraietta JA, Davis MM, Cohen AD, Weber KL, Lancaster E, Mangan PA, Kulikovskaya I, Gupta M, Chen F, Tian L, Gonzalez VE, Xu J, Jung IY, Melenhorst JJ, Plesa G, Shea J, Matlawski T, Cervini A, Gaymon AL, Desjardins S, Lamontagne A, Salas-Mckee J, Fesnak A, Siegel DL, Levine BL, Jadlowsky JK, Young RM, Chew A, Hwang WT, Hexner EO, Carreno BM, Nobles CL, Bushman FD, Parker KR, Qi Y, Satpathy AT, Chang HY, Zhao Y, Lacey SF, June CH. CRISPR-engineered T cells in patients with refractory cancer. Science. 2020 Feb 28;367(6481):eaba7365. doi: 10.1126/science.aba7365. Epub 2020 Feb 6. PMID: 32029687.
- Ebina H, Misawa N, Kanemura Y, Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep. 2013;3:2510. doi: 10.1038/srep02510. PMID: 23974631; PMCID: PMC3752613.
- Liao HK, Gu Y, Diaz A, Marlett J, Takahashi Y, Li M, Suzuki K, Xu R, Hishida T, Chang CJ, Esteban CR, Young J, Izpisua Belmonte JC. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat Commun. 2015 Mar 10;6:6413. doi: 10.1038/ncomms7413. PMID: 25752527.
- Kaminski R, Chen Y, Fischer T, Tedaldi E, Napoli A, Zhang Y, Karn J, Hu W, Khalili K. Elimination of HIV-1 Genomes from Human T-lymphoid Cells by CRISPR/Cas9 Gene Editing. Sci Rep. 2016 Mar 4;6:22555. doi: 10.1038/srep22555. Erratum in: Sci Rep. 2016 Jul 07;6:28213. PMID: 26939770; PMCID: PMC4778041.
- Herrera-Carrillo E, Berkhout B. Attacking HIV-1 RNA versus DNA by sequence-specific approaches: RNAi versus CRISPR-Cas. Biochem Soc Trans. 2016 Oct 15;44(5):1355-1365. doi: 10.1042/BST20160060. PMID: 27911718.
- Lebbink RJ, de Jong DC, Wolters F, Kruse EM, van Ham PM, Wiertz EJ, Nijhuis M. A combinational CRISPR/Cas9 gene-editing approach can halt HIV replication and prevent viral escape. Sci Rep. 2017 Feb 8;7:41968. doi: 10.1038/srep41968. PMID: 28176813; PMCID: PMC5296774.
- Xu L, Wang J, Liu Y, Xie L, Su B, Mou D, Wang L, Liu T, Wang X, Zhang B, Zhao L, Hu L, Ning H, Zhang Y, Deng K, Liu L, Lu X, Zhang T, Xu J, Li C, Wu H, Deng H, Chen H. CRISPR-Edited Stem Cells in a Patient with HIV and Acute Lymphocytic Leukemia. N Engl J Med. 2019 Sep 26;381(13):1240-1247. doi: 10.1056/NEJMoa1817426. Epub 2019 Sep 11. PMID: 31509667.
- Yin L, Zhao F, Sun H, Wang Z, Huang Y, Zhu W, Xu F, Mei S, Liu X, Zhang D, Wei L, Cen S, Hu S, Liang C, Guo F. CRISPR-Cas13a Inhibits HIV-1 Infection. Mol Ther Nucleic Acids. 2020 Sep 4;21:147-155. doi: 10.1016/j.omtn.2020.05.030. Epub 2020 Jun 1. PMID: 32585623; PMCID: PMC7321785.
- Wilkie T, Panfil AR. CRISPR Targeting the Integrated HTLV-1 Virus. Biotechnologies for Gene Therapy. 2022. SpringerLink. https://link.springer.com/chapter/10.1007/978-3-030-93333-3_6
- Tanaka A, Takeda S, Kariya R, Matsuda K, Urano E, Okada S, Komano J. A novel therapeutic molecule against HTLV-1 infection targeting provirus. Leukemia. 2013 Aug;27(8):1621-7. doi: 10.1038/leu.2013.46. Epub 2013 Feb 15. PMID: 23411465.
- Nakagawa M, Shaffer AL 3rd, Ceribelli M, Zhang M, Wright G, Huang DW, Xiao W, Powell J, Petrus MN, Yang Y, Phelan JD, Kohlhammer H, Dubois SP, Yoo HM, Bachy E, Webster DE, Yang Y, Xu W, Yu X, Zhao H, Bryant BR, Shimono J, Ishio T, Maeda M, Green PL, Waldmann TA, Staudt LM. Targeting the HTLV-I-Regulated BATF3/IRF4 Transcriptional Network in Adult T Cell Leukemia/Lymphoma. Cancer Cell. 2018 Aug 13;34(2):286-297.e10. doi: 10.1016/j.ccell.2018.06.014. Epub 2018 Jul 26. PMID: 30057145; PMCID: PMC8078141.
- Laydon DJ, Sunkara V, Boelen L, Bangham CRM, Asquith B. The relative contributions of infectious and mitotic spread to HTLV-1 persistence. PLoS Comput Biol. 2020 Sep 17;16(9):e1007470. doi: 10.1371/journal.pcbi.1007470. PMID: 32941445; PMCID: PMC7524007.
- Mout R, Ray M, Lee YW, Scaletti F, Rotello VM. in vivo Delivery of CRISPR/Cas9 for Therapeutic Gene Editing: Progress and Challenges. Bioconjug Chem. 2017 Apr 19;28(4):880-884. doi: 10.1021/acs.bioconjchem.7b00057. Epub 2017 Mar 17. PMID: 28263568; PMCID: PMC5846329.

Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
