Journal of Clinical Microbiology and Biochemical Technology
Microbial enzymatic activity measurements by fluorogenic substrates: Evidence of inducible enzymes in oligotrophic Mediterranean areas
Caruso G1*, Caruso R2 and Maimone G1
2Hospital Agency A.O.U. “G. Martino”, Messina, Italy
Cite this as
Caruso G, Caruso R, Maimone G (2019) Microbial enzymatic activity measurements by fluorogenic substrates: Evidence of inducible enzymes in oligotrophic Mediterranean areas. J Clin Microbiol Biochem Technol 5(2): 019-024. DOI: 10.17352/jcmbt.000034Background: In aquatic environments, organic polymers such as proteins, polysaccharides and organic phosphates are cleaved and up taken by microorganisms through the expression of specific enzymes such as Leucine Aminopeptidase (LAP), beta Glucosidase (GLU) and Alkaline Phosphatase (AP), respectively. Microbial enzymatic activities are a fundamental step in the organic matter utilization and turnover into simple monomers.
The context and purpose of the study: An experiment was carried out by combining the enzymatic assay using fluorogenic substrates with microscopical counts obtained using the viability marker 5-cyano-2,3 ditolyl-tetrazolium chloride (CTC). The main objective was to verify whether the enzyme activity played by microbial cells depends of the cell viability or rather whether the hydrolytic activity is directly stimulated by the availability of organic substrates for microbial metabolism.
Results: This study provides evidence that in oligotrophic waters the fluorescence signal often decreases after incubation with the substrate analogue used for the enzymatic assay. For such samples, the extension of the incubation period from 2 to 5 hours has allowed the detection of a positive fluorescence signal. Simultaneous counts of the abundance of CTC+ cells revealed that only a fraction close to 30% of the total bacterioplankton was actively respiring, suggesting that the observed increase was related to the presence of inducible enzymes rather than of actively metabolising cells.
Main findings: Since the synthesis of hydrolytic enzymes by the microbial community can be induced by the presence of the organic substrates, adapting the incubation period to the trophic condition of the examined area is required for accurate enzymatic measurements, especially for oligotrophic environments.
Conclusions: This is the first contribution to link the enzyme activity rates with the viability properties (in terms of actively metabolising components) of bacterioplankton inhabiting pelagic Mediterranean waters.
Brief summary: Evidence of inducible enzymes in oligotrophic Mediterranean areas was provided by the increase of fluorescence recorded after incubation up to 5 hours with specific fluorogenic substrates, in concomitance with the detection of a low fraction of actively respiring cells. While microbial cells were quite active for AP synthesis, LAP and GLU were mostly inducible enzymes, activated by the addition of their specific polymeric organic substrate.
Any potential implications: In the examined oligotrophic waters the enzymatic activities seem to be stimulated mostly by the availability of metabolisable organic substrates.
Introduction
Microorganisms play a fundamental role in the functioning of the marine ecosystem and particularly in the transfer of energy at the basis of the trophic web. The different size (pico-, nano and micro-plankton) and functional planktonic classes (bacterio-, phyto- and zoo-plankton) perform autotrophic and heterotrophic activities that drive marine biogeochemical processes. Knowledge of the specific role played by different planktonic size organisms and of their biogeochemical implications in different ecological contexts attracts increasing attention by the scientific community interested in marine ecosystem health and safeguard to get accurate environmental assessment in management programs [1-4]. Understanding all the biodiversity components (among which microbes) that regulate the functioning of the marine environment represents one of the most intriguing research challenges for marine ecologists [5]. To this end, linking microbial enzymatic activity measurements with the assessment of the metabolic conditions of the prokaryotic community is of great significance to better elucidate the microbial role in biogeochemical cycles.
In the Mediterranean Sea preservation of the health status of this ecosystem represents a current strategic objective under the imperative indications of the Marine Strategy Framework Directive [6]; in this context, inclusion of microorganisms -until now neglected by the European Commission Decision 2010/477 on criteria and methodological standards related to the Good Environmental Status-GES (Table 1 of Annex III)- as a key component in decision-making contexts and for the best protocol design practices of sea monitoring has been indicated as a relevant knowledge gap by several studies [3,4,7]. On the other hand, implementation of observational and experimental data concerning the quantification of organic carbon transformation and remineralization along the water column mediated by the biological and microbial carbon pumps, is also the subject of European research programmes like MERMEX, SESAME (European Commission_FP6), PERSEUS (European Commission_FP7).
Previous estimates of microbial enzymatic activities performed in a range of marine environments [3,8,9] have highlighted that in oligotrophic waters frequently the measurements taken after the addition of the fluorogenic substrates did not yield a detectable fluorescence signal; the readings recorded after incubation with the reagent were often lower than the initial measurements. This observation suggested that the added substrate was not immediately metabolised by the microbial community and that the enzyme needed for substrate hydrolysis was induced only after incubation with its specific substrate (called “inducible enzyme”). In other words, the time delay observed until a positive fluorescent signal was measured suggested that the enzymatic synthesis was regulated by the relative richness in organic matter of the sample. Therefore, in pelagic environments, characterised by oligotrophic conditions, the microbial community was expected to be equipped with inducible enzymes, while in eutrophic environments such as coastal ones, enzymes are directly expressed, without a stimulation by their specific substrate (called “constitutive” enzymes).
An experiment was undertaken to verify the following working hypothesis: is the enzyme activity related to cell viability as a marker of cell physiological state, or rather is microbial metabolism directly stimulated by the presence of organic polymers? In this experiment the enzymatic degradation of proteins, carbohydrates and organic phosphates at pelagic marine stations was studied through measures of the specific enzymes Leucine Aminopeptidase (LAP), beta-Glucosidase (GLU) and Alkaline Phosphatase (AP), respectively. To relate the expression of enzymatic activity to the physiological state of microbial cells, the enzymatic assays were combined with estimates of cell viability using CTC stain as a marker of actively respiring cells.
Materials and Methods
Collection and treatment of seawater samples
Within the monitoring plans prescribed by the Italian Ministry for Environment to meet the indications of the Marine Strategy Framework Directive (Directive 2008/ 56EC), the cruise “Marine Strategy Adriatic- Ionian Sea” was performed in September 2016 in two Assessment Areas including the Central Ionian Sea and the Southern Adriatic basin.
A total of 10 pelagic stations were sampled during the cruise (Figure 1) at three depths of the water column, identified as Surface, Deep Chlorophyll Maximum- DCM (corresponding to the peak of chlorophyll-a) and Deep layer, corresponding to the lowest depth of the photic layer (generally around 100 m). The collected water samples were stored onboard at +5°C until their analysis, within maximum 3 days of sampling. At the arrival in laboratory, they were treated differently according to the parameters to be determined.
Enzymatic activity rates
Enzymatic activity measurements were performed on unfiltered bulk water samples using the Hoppe’s method [10], based on the cleavage of fluorogenic substrates derived from Methylcoumarine (MCA) or Methylumbellyferone (MUF). The substrates used for this study were: L-leucine-7-amido-4-methylcoumarin hydrochloride (leucine-MCA), 4-Methylumbelliferyl-β-D-Glucopyranoside (MUF-β-glucose), and Methylumbelliferyl-Phosphate (MUF-phosphate), that are specific for the enzymes Leucine Aminopeptidase (LAP), beta-Glucosidase (GLU) and Alkaline Phosphatase (AP), respectively. They were added to 10 ml- subvolumes of samples, at increasing concentrations ranging from 20 to 160 micromoles per litre, according to the standard laboratory procedure described by Caruso [1]. The potential rates of hydrolysis of the substrates were determined with a Glomax fluorimeter using the UV filter for excitation and recording the Raw Fluorescence Units (RFU) of the fluorescence signal released after the cleavage of the substrate. Measurements were taken at time T1, immediately after the addition of the substrate, and at successive times (T2 and T3, after 2 and 5 hours of incubation at “in situ” temperature). The RFU were further referred to a standard curve obtained with known concentrations of the standards MCA and MUF to calculate the potential activity of each enzyme.
Cell viability estimates using CTC labelling
To detect the fraction of actively respiring cells, samples collected from 4 stations at 3 depths (for a total of 12 samples) were selected as representative of different sub-basins, particularly station 22, located in the Sicily Channel; station 66, in the Calabrian area of the Ionian Sea; station 96, in the crossing area between the Ionian and the Southern Adriatic Seas, and station 112, in the Apulian area of the Southern Adriatic Sea.
Before microscopical counts, water sub-samples (10ml) were pre-filtered through a 100µm-mesh size net to prevent clogging of the pores of the filtering membrane for epifluorescence microscopy. CTC is a redox stain that in presence of oxygen is converted by electron transport system into formazan, which accumulates as red fluorescing granules within the actively respiring cells (CTC+ cells) [11]. Aliquots (2ml) of the filtered sample were stained with addition of a 5mmol l-1 solution of CTC (final concentration; Polyscience. Warrington. Pa), freshly prepared from the stock solution obtained according to manufacturers’ instructions. After incubation for 5 hours with CTC, the treated samples were filtered through a Nuclepore black (25mm diameter, 0.22µm pore size) polycarbonate filter; the filter was further mounted on a glass slide using a drop of Immersoil 158F (Zeiss).
Microscopical analysis
Microscopical counts were made on at least 20 fields with a Zeiss Axioplan 2 epifluorescence microscope (Carl Zeiss Vision GmbH, Munchen, Germany) coupled with an image analysis system, equipped with a 100W mercury lamp and specific filter sets (green light: BP 510-560, FT 580 and LP 590). Observations were performed using a Neofluar objective at a 1000X magnification under immersion oil; CTC+ cells were viewed as red fluorescing cells due to intracellular formazan granules and their counts were expressed as the mean value of cells observed per ml of sample (cells ml-1).
Statistical analysis
Statistical relationships existing between the increase of fluorescence and the concentration of CTC+ cells were analysed by Pearson correlation; only the values reaching a significance level of 0.05% of probability were considered as statistically significant.
Results
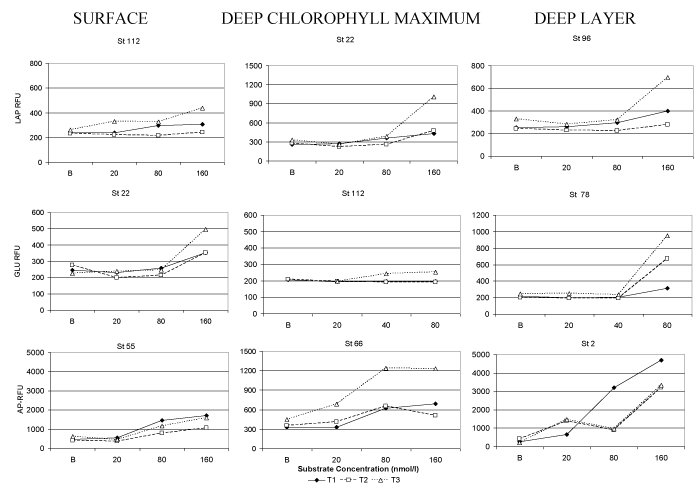
The typical course depicted by the measurements of fluorescence intensity in relation to the amount of added substrate is shown in Figure 2. The percentage of fluorescence increase measured using our modified protocol of enzymatic assay is shown in Table 1, separately for the three depths of water column (Surface, DCM and Deep layer). The greatest increases in fluorescence were generally recorded at substrate concentrations of 160nmol/l for LAP and AP, and also of 80nmol/l for GLU; these concentrations were considered to be sufficient enough to induce enzymatic production. For AP, fluorescence measurements gave for most of the samples positive values at time T2 compared to T1, and increases in fluorescence readings were recorded mainly at stations 22, 32, 55 and 66 and in the samples taken from the bottom depths of the other stations. For GLU, increases in fluorescence after 2 hours of incubation were observed only at stations 2 and 10, while for LAP activity, all the fluorescence readings performed at time T2 were lower than those recorded at time T1. With the modified analytical protocol, the rates of fluorescence increase (as an index of the recovery of metabolic activity) varied from a minimum of 5.7 (st. 55, deep layer) to a maximum of 99.5% of the value initially recorded ( st. 66, surface) for LAP, from 1.5 (st. 32- DCM) to 40.1% (stat. 55, surface) for AP and from 2.4 (stat 55, deep layer) to 77.1% (stat 78, surface) for GLU.
Along the vertical water column, the microbial metabolic rates showed a greater increase of fluorescence signal at DCM compared to the deepest layer.
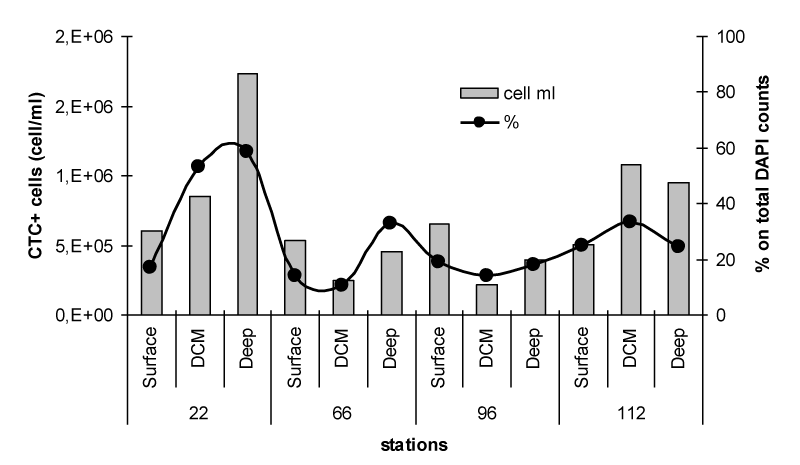
The pattern of spatial distribution of actively respiring bacterial cells, determined as the abundance of CTC labelled bacteria, is reported in Figure 3. A different predominance of actively metabolising cells among the examined samples was observed, with the highest counts at station 22 (deepest layer) and 112 (DCM and deepest layer). At these depths, the active cells accounted for a percentage close to 30% of total bacterioplankton, reaching a peak of 53% at the Deep layer of station 22. Conversely, low abundance of CTC+ cells was found at the stations 66 and 96, especially in correspondence of the DCM.
The outputs of Pearson’s correlation analysis are reported in Table 2. Higher, although not statistically significant, negative relationships were found between the abundance of CTC+ cells and the increase of enzymatic activity computed between T3 and T2. The negative relationships calculated between CTC+ cell counts and the increase in fluorescence readings obtained with the modified enzymatic protocol probably depended on the active metabolism shown by the microbial community of the analysed samples already at T2, such as in the case of AP; for this reason enzyme activity did not increase further when the incubation time was prolonged to T3. This inverse result could also suggest that the increase of the hydrolytic activity did not depend on the physiological state of the bacterial cells, as the low counts of actively respiring cells apparently suggested; rather, it could depend on the presence of inducible enzymes, whose expression was stimulated by the availability of organic matter (in this case the added substrate) that could be metabolised.
Discussion
In aquatic microbial ecology, the rates of microbial enzymatic activity provide indications on the presence of a microbial community with active metabolic capacities and capable of ensuring the degradation of organic matter and the recycle of nutrients to sustain the functioning of the entire ecosystem. Therefore, microbial enzymatic activity measurement can contribute, together with other physical-chemical and biological parameters, to the formulation of a more complete judgment on the environmental quality. It is commonly expected that in natural oligotrophic environments microbes are adapted to live under limited resource availability, therefore amounts of organic substrates exceeding the normal microbial metabolic capability (such as those from the fluorogenic substrate even if experimentally added at very low concentrations) are not immediately metabolised, but after a certain latency period. Indeed, increases in the proportion of active cells, and decreases in that of inactive or dormant cells, are usually reported along gradients of increasing system productivity [12]. On the other hand, being microbial cells able to survive “in situ” even in conditions of resource limitation (such as those typical of oligotrophic environments), it is likely that their cell physiological state can affect their metabolic response to the availability of organic matter.
In the present study, a first experiment was performed in the Mediterranean basin, where enzymatic data have been related to viability parameters. The activity rates of LAP, GLU and AP were analysed and compared with the abundance of actively respiring bacteria, as determined by epifluorescence microscopical count of the cells labelled with the fluorochrome CTC, in order to relate the abundance of living cells to their role in the ecosystem functioning. The potential enzymatic activity values estimated in the examined samples (data not shown, varying from 0.09 and 4.788nmol/l/h for LAP, 0.113 to 11.23nmol/l/h for GLU, and 0.378 to 200.52nmol/l/h for AP) were generally comparable in magnitude order with those reported in other pelagic Mediterranean areas [3,8,9,13]. Comparison of the activity levels obtained in the present survey with previous data measured in the same basin was the only possible, since no reference values for enzymatic activity rates are currently reported by the Legislation.
At a first glance, the decreasing trend of fluorescence readings in the examined oligotrophic water samples recorded after 2 hours of incubation with the fluorogenic substrate could be explained by hypothesizing that most of microbial community consisted of stressed or dormant cells. In natural aquatic environments, microbes are affected by several factors, such as solar radiation, temperature, salinity, water pressure, resource availability, grazing pressure. Microorganisms exposed to adverse environmental factors may also adopt a viable but nonculturable survival strategy as a strategy to cope with them [14] and cell dormancy is a very widespread physiological condition in aquatic ecosystems [15]. Indeed, it is well known that viable culturable bacteria account for a very small fraction of total bacterioplankton, i.e. less than 1% in ocean waters [16] and from 0.001 to 0.2% in pelagic waters of the north-western Mediterranean Sea [17]) and that within natural aquatic communities only a low fraction (on average, 30% of total abundance) of bacteria is active [18]. The mean proportion of CTC+ cells found in the present study was consistent with such estimates. In ecological studies, the quantification of the bacterial fraction that is still alive is essential to correlate the abundance of active cells with the activity they effectively play within the biological processes [19-20]. In our study case, however, the measure of discrete activity rates of AP led us to think the microbial cells were physiologically active and not dormant cells. Comparing the reciprocal patterns of the three enzymatic activities, a lower percentage of fluorescence increase was recorded for AP measurements, suggesting that this enzyme was already expressed by the microbial community and that incubation for 2 hours was sufficient enough to detect a positive fluorescence signal. This result could be related to the fact that AP is an enzyme produced not only by bacteria but also by phytoplankton, and therefore AP activity levels are generally well above the detection limits. In synthesis, in the examined oligotrophic environments enzymatic measurements required extending the incubation time from 2 to 5 hours for the full development of the hydrolytic activity. This lag phase required for the detection of enzyme activity was observed especially for the enzymes LAP and GLU, while AP seemed to be already expressed by the microbial community of the examined environments, acting as a constitutive enzyme. Conversely, AP was previously described [21] as an inducible catabolic enzyme, whose synthesis is regulated by the concentration of the inorganic phosphorous. The presence of two distinct-inducible and constitutive-AP was previously documented in Serratia marcescens cultured at low phosphate concentrations [22]. Previous observations have documented that, like most other hydrolytic ectoenzymes of aquatic microorganisms, β-Glucosidases are a heterogeneous group of hydrolytic enzymes produced by yeasts, fungi, and bacteria; they are inducible enzymes involved in the induction of cellulase and in the degradation of cellulose [23].
Compared with reports on enzyme activity measurements currently adopted in aquatic microbial ecology [1,10], in the present study the combination of the enzyme assay with the viable staining protocol gave us the advantage to estimate whether and to what extent microbial cells were metabolically active and to associate this information with the fraction of the whole microbial community retaining viable attributes, as shown by the percentage of labelling with the CTC viability stain.
Since the actual percentage of active prokaryotic cells and the rates of the microbial processes related to biogeochemical cycles is still largely unknown, the obtained findings underline that further studies on the links between microbial community viability and activity as well as on their driving environmental factors are recommended to improve current knowledge of the ecological relevance of microbial community role and comprehension of aquatic ecosystems functioning.
The Authors thank the Captain, the crew of RV Minerva Uno and the colleagues dr. Maurizio Azzaro (CNR-ISP, Messina) and Mr. Giuseppe Arena (University of Messina) for their help with sampling during the “Marine Strategy- Adriatic/Ionian Sea” oceanographic cruise. Funds supporting this research were provided by CNR, within the Italian Flag Project Ritmare and the Italian Marine Strategy Framework Directive Programmes.
- Caruso G (2010) Leucine aminopeptidase, beta-glucosidase and alkaline phosphatase activity rates and their significance in Carbon and Phosphorus cycles in some coastal Mediterranean sites. Mar Drugs 8: 916-940. DOI: 10.3390/md8040916. Link: http://bit.ly/2NOOPdS
- Caruso G, Decembrini F, Caruso R, Zappalà G, Bergamasco A, et al. (2011) Are microbial enzyme activities sensitive indicators of the trophic state of marine ecosystems? In Pollution Monitoring, Edited by Ortiz AC and Griffin NB. Hauppauge NY, USA: NOVA 195-210. Link: http://bit.ly/2NNP6xp
- Caruso G, La Ferla R, Azzaro M, Zoppini A, Marino G, et al. (2016) Microbial assemblages for environmental quality assessment: knowledge, gaps and usefulness in the European Marine Strategy Framework Directive. Crit Rev Microbiol 42: 883-904. Link: http://bit.ly/2rHuBd7
- Garuso G, Azzaro M, Caroppo C, Decembrini F, Monticelli LS, et al. (2016) Microbial community and its potential as descriptor of environmental status. ICES J Mar Sci 73: 2174–2177. Link: http://bit.ly/32PTsIO
- Borja A (2014) Grand challenges in marine ecosystems ecology. Front Mar Sci 1: 1-6. Link: http://bit.ly/2Okr78m
- European Commission (2008) Directive 2008/56/EC of the European Parliament and of the Council of the 17 June 2008 establishing a framework for Community actions in the field of marine environmental policy (Marine Strategy Framework Directive). Official J Eur Comm L164:19-40. Link: http://bit.ly/2potnCY
- Caroppo C, Buttino I, Camatti E, Caruso G, De Angelis R, et al. (2013) State of the art and perspectives on the use of planktonic communities as indicators of environmental status in relation to the EU Marine Strategy Framework Directive. Biol Mar Medit 20: 65-73. Link: http://bit.ly/2CHdNp4
- Zaccone R, Caruso G, Azzaro M, Azzaro F, Crisafi E, et al. (2010) Prokaryotic activities and abundance in pelagic areas of the Ionian Sea. Chem Ecol 26: 169-197. Link: http://bit.ly/32NTCjJ
- La Ferla R, Azzaro M, Caruso G, Monticelli LS, Maimone G, et al. (2010) Prokaryotic abundance and heterotrophic metabolism in the deep Mediterranean Sea. Adv Oceanogr Limnol 1: 143-166. Link: http://bit.ly/2XhsrwP
- Hoppe HG (1993) Use of fluorogenic model substrates for extracellular enzyme activity (EEA) measurement of bacteria. In Handbook of methods in aquatic microbial ecology, Edited by Kemp PF, Sherr BF, Sherr EB, Cole JJ. Boca Raton, FL, USA: Lewis Publisher: 423-432. Link: http://bit.ly/377giie
- Sherr BF, del Giorgio P, Sherr EB (1999) Estimating abundance and single-cell characteristics of respiring bacteria via the redox dye CTC. Aquat Microb Ecol 18: 117-131. Link: http://bit.ly/2Krejfc
- Kirchman DL (2008) Microbial ecology of the oceans- 2nd ed. Wiley & Sons, Inc, Hoboken, New Jersey. Link: http://bit.ly/3418igI
- Placenti F, Azzaro M, Artale V, La Ferla R, Caruso G, et al. (2018) Biogeochemical patterns and microbial processes in the Eastern Mediterranean Deep Water of Ionian Sea. Hydrobiologia 815: 97-112. Link: http://bit.ly/2KDqwh1
- Joux F, Lebaron P (2000) Use of fluorescent probes to assess physiological functions of bacteria at single-cell level. Microb Infect 2: 1523-1535. Link: http://bit.ly/374lN1g
- Lennon JT, Jones SE (2011) Microbial seed banks: the ecological and evolutionary implications of dormancy. Nature Rev Microbiol 9: 119-130. Link: https://go.nature.com/2Kp3ief
- Kogure K, Simidu U, Taga N (1979) A tentative direct microscopic method for counting living marine bacteria. Can J Microbiol 25: 415-420. Link: http://bit.ly/2CMAQi6
- Bianchi A, Giuliano L (1996) Enumeration of viable bacteria in the marine pelagic environment. Appl Environ Microbiol 62: 174-177. Link: http://bit.ly/2KoIsf4
- Smith EM, del Giorgio PA (2003) Low fractions of active bacteria in natural aquatic communities. Aquat Microb Ecol 31: 203-208. Link: http://bit.ly/2KmgUqs
- Grégori G, Citterio S, Ghiani A, Labra M, Sgorbati S, et al. (2001) Resolution of viable and membrane-compromised bacteria in freshwater and marine waters based on analytical flow cytometry and nucleic acid double staining. Appl Environ Microbiol 67: 4662-4670. Link: http://bit.ly/354IHUi
- Caruso G, Zappalà G Maimone G, Azzaro F, Raffa F, et al. (2008) Assessment of the abundance of actively respiring cells and dead cells within the total bacterioplankton of the Strait of Messina waters. WIT Transact on Built Environ 99: 15-24. Link: http://bit.ly/2rNWyAf
- Hoppe HG (2003) Phosphatase activity in the sea. Hydrobiologia 493: 187-200. Link: http://bit.ly/2CMBA6S
- Bhatti AR, Alvi A, Chaudhry GR (2000) Evidence on the presence of two distinct alkaline phosphatases in Serratia marcescens. FEMS Microbiol Lett 182: 131-135. Link: http://bit.ly/2XhFbTO
- Zang X, Liu M, Fan Y, Xu J, Xu X, et al. (2018) The structural and functional contributions of β-glucosidase-producing microbial communities to cellulose degradation in composting. Biotechnol Biofuels 11: 51. Link: http://bit.ly/2Odeg7Q

Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
