Journal of Biology and Medicine
Different Strategies used in the Production of human monoclonal scfv Antibodies Specific to Dimers of Membrane Receptors
Sylwia Łukasiewicz*, Agata Stachowicz, Ewelina Fic, Ewa Błasiak, Agata Kowalik and Marta Dziedzicka-Wasylewska
Cite this as
Łukasiewicz S, Stachowicz A, Fic E, Błasiak E, Kowalik A, et al. (2019) Different Strategies used in the Production of human monoclonal scfv Antibodies Specific to Dimers of Membrane Receptors. J Biol Med 3(1): 014-020. DOI: 10.17352/jbm.000007Phage display technology is one of the methods widely adopted for the production of human monoclonal antibodies. The subject of the present research was the scFv - single chain variable fragment antibody - which was able to recognize the D2-5-HT1A heteromer formed by dopamine D2 and serotonin 5-HT1A receptors. The antibody was selected using the phage display technique and the human antibody scFv phagemid library Tomlinson J. We investigated two expression systems: prokaryotic and eukaryotic. The results indicate that both of these systems (E.coli HB2151 and High Five cells (BTI-TN-5B1-4) of Trichopulsia ni) are suitable for the production of scFv antibodies, but the scFvs obtained from insect cells could attain a higher protein concentration. Next, we examined a few purification methods: immobilized-metal affinity chromatography (IMAC), affinity chromatography with the c-myc tag and the methods using protein L or A agarose, which are applicable to scFvs lacking tags. In the bacterial as well as insect cell expression systems, protein L agarose chromatography gave satisfactory results, and in our opinion, it is the best choice as a purification resin. Additionally, circular dichroism measurements confirmed the predominant content of secondary structures in the tested antibodies, which is consistent with the literature data.
Introduction
ScFvs (single chain variable fragment) are small antibody fragments composed of immunoglobulin-heavy (VH) and light chain-variable (VL) regions with a flexible peptide linker designed to connect the two chains such that the antigen binding site is retained in a single co-linear molecule [1]. Currently scFvs antibodies attract a lot of attention due to their potential usage in pharmacology and diagnostic fields [2-4]. The properties of this kind of antibodies make them an attractive modality for pharmaceutical development, especially in areas such as cancer and autoimmune diseases in which cell-cell interactions or ligand-receptor interactions play a crucial role in the pathogenesis [5]. Moreover, scFvs are very useful nanoprobes for the targeted delivery of drugs to specific cells or tissues since they can be successfully used as targeting ligands for the functionalization of nanocarriers [6]. In comparison to the much larger Fab, F(ab)2, and IgG forms of the monoclonal antibodies from which they are derived, scFvs have lower retention times in non-target tissues, more rapid blood clearance, better tissue penetration and reduced immunogenicity, making them attractive for therapeutic applications [7].
ScFvs antibodies can be derived from phage display libraries [8,9]. Phage display technology (based on the significant work of Smith, 1985) [10] is an in vitro screening technique that allows rapid selection of multiple monoclonal antibodies of high affinity and specificity [11]. The nature of the method enables the rapid and efficient identification of specific binders, as well as greater control over the selection parameters that facilitate the isolation of antibodies with unique, desirable functional characteristics [5].
Very often, scFvs contain poly-His or/and c-myc tags that are used in their purification and identification in vitro. Many protocols describe scFv production and purification in specific expression systems, including bacteria, yeast, plant, insect, and mammalian cells, as well as in transgenic animals [12-15]. Depending on the expression system, their ability to fold and secrete the scFv proteins varies [16].
In this report, we investigated two expression systems: prokaryotic and eukaryotic. We present the results from the preparation of scFv expressed in E.coli HB2151 and High Five cells (BTI-TN-5B1-4) of Trichopulsia ni. Prokaryotic expression systems have several advantages including low cost of production and the ability to produce large amounts of recombinant proteins in short periods of time, but their main drawback is the accumulation of aggregated material in the cytoplasm, known as inclusion bodies. On the other hand, a significant advantage of insect cells is their capability to perform most posttranslational modifications used by higher eukaryotes, such as glycosylation, phosphorylation, acylation and palmitylation [17]. In the present work, we focused on developing a simple, fast and efficient method for the purification of soluble scFv fragments. We examined a few purification methods: immobilized-metal affinity chromatography (IMAC), which is only applicable to scFvs containing a poly His-tag; affinity chromatography with the c-myc tag; and the methods using protein L or A agarose, which are applicable to scFvs lacking tags. The subject of the research was scFvs antibodies specifically recognizing D2-5-HT1A heteromer (dimer formed by dopamine D2 and serotonin 5-HT1A receptors) selected using the phage display technique and the human antibody scFv phagemid library Tomlinson J. The presented results have been collected for at least several various scFv proteins whose common characteristic was selectivity against D2-5-HT1A membrane proteins. Although we described data only for different types of scFvs antibodies selective for one type of heteromer, the presented results were similar also for other types of scFvs antibodies obtained against other various membrane receptors (data not shown). We choose scFvs against D2-5-HT1A heteromer for publication as a representative case since it would be difficult to show results for all proteins which were obtained in our lab and it would make the work unreadable and confusing. Bearing in mind difficulties in obtaining scFv specifically recognizing dimers of membrane receptors the statistical attempt and data collected in the presented work seem to be interesting and may be a good starting point in designing similar experiments.
Materials and Methods
Selection of human monoclonal scFv antibody
Human monoclonal scFvs antibodies were selected using the phage displayed library Tomilson J (Geneservise) which was amplified and titrated according to the manufacturer’s protocols using E. coli TG1 cells. In total, 1.9x1013 individual clones were panned and screened against D2-5-HT1A heteromers presented on the surface of CHO-K1 cells stably expressing both desired receptors. Five rounds of positive selection followed by negative preselection (where CHO-K1 cells expressing D2R mixed with CHO-K1 cells expressing 5-HT1AR (1:1) (CHO-negative cells; CHO-) were used) and finally 2 negative selections were performed. Before protein expression, the cDNAs of scFvs from the selected phages were isolated and sequenced to check for the presence of STOP codons inside the coding sequence.
Prokaryotic expression system
To obtain soluble scFv expressed in E. coli, HB2151 (Novagen) cells were transformed with selected vector plasmids. A single colony was used to inoculate 5 ml of 2xYT medium+Amp (100 μg/ml) (Lab Empire), and the cultures were grown overnight at 37°C. A 0.5 ml portion of the overnight culture was used to inoculate 25 ml of medium, and then this culture was used to start a 1L culture at 37°C. Expression was induced in the host bacteria by adding IPTG (Sigma-Aldrich) to a final concentration of 1 mM when the OD600 reached 0.9. Cells were cultured for 16 h at 28°C, and then the cells were harvested by centrifugation (4000 g, 15 min, 4°C). The cells were placed on ice until the purification of scFv procedure was started.
Eukaryotic expression system
The production of scFv in eukaryotic cells was conducted with the Bac-to-Bac Baculovirus Expression System from Invitrogen (Thermo Fisher Scientific). Sf9 and High Five cells were cultured in ESF 921 Insect Cell Culture Medium from Expression Systems supplemented with antibiotic-antimycotic solution (Sigma-Aldrich). Cells were maintained and expanded at 28°C without CO2 as monolayer cultures in tissue culture bottles or as suspension cultures in Erlenmeyer shake flasks.
The cDNAs of the selected scFv antibodies were cloned using EcoRI and HindIII into the pFastBac1 vector (Bac-to-Bac Baculovirus Expression System, Invitrogen). Double digestion with the same restriction enzymes was used to confirm the correct cDNA subcloning. Recombinant bacmids were generated by Tnt transposition into DH10Bac E. coli cells (Invitrogen). In this procedure, 1 mg of the donor plasmids (pFastBac) containing the scFv antibody cDNAs was used for transformation into the DH10Bac cells. The transformed DH10Bac bacteria were grown for 48 h at 37°C on selective LB agar plates containing 7 mg/ml gentamicin, 10 mg/ml tetracycline, 50 mg/ml kanamycin, 100 mg/ml Bluo-gal and 40 mg/ml IPTG. Recombinant bacmid DNAs were extracted from white single colonies obtained in a blue-white test. Transposition of the scFv cDNA into the bacmid DNA was confirmed with PCR. Positively verified recombinant bacmids were used for baculovirus production. Sf9 cells were transfected with 3 μg of recombinant bacmids using Cellfectin-II (Invitrogen). Transfected cells were allowed to grow in a monolayer culture and were observed daily for signs of viral infection and cytopathic effects. At 72 hours after transfection, baculoviruses were harvested (200 g, 10 min, 4°C), and baculovirus infectious titers were determined in a viral plaque assay. Baculoviruses were amplified in Sf9 cells and then used to infect High Five cells with a multiplicity of infection 5. Cells were harvested 48 hours after infection (200 g, 10 min, 4°C) and stored at -80°C until the purification steps.
Expression level analysis
After scFv expression obtained periplasmatic fraction from E.coli HB2151 as well as cell lisates from High Five cells were analysed by ELISA technique using affinity of the scFv (Tomilson J) fragments to protein A. Protein A resuspended in PBS buffer (1 μg/ml) was added to each well of 96-well plate (50 μl per well) and incubated overnight at 4°C. Then the plate was blocked using 2 % BSA solution (1 h, 37°C) and 50 μl of investigated samples (periplasmatic fraction or cell lysates) were added to appropriate wells and incubated at 37°C for 1 h. The washing step was conducted 3 times 200 ml PBS. Then 50 ml of protein L conjugated with HRP (protein L was resuspended in 0.5% BSA/PBS at a 1:2000 ratio) (GE Healthcare) was added to the wells and incubated for 30 min at 37°C. After washing (carried out 4 times as described above), the reactions were developed by TMB substrate (GE Healthcare). Experiments were performed in triplicate.
scFv purification
ScFvs derived from E.coli and insect cells were purified at room temperature, but the buffers used were ice-cold.
Bacterial expression system: ScFvs are secreted into the periplasmic space located between the outer and inner membranes of bacteria. The pellet of bacterial cells was resuspended in buffer consisting of 50 mM Tris-HCl, 20% sucrose, pH 8.0, 1 mM EDTA, and 1x Protease Inhibitor Cocktail (20 ml buffer/1L overnight culture). The cells were incubated on ice for 15 min with gentle agitation, then the cell suspension was centrifuged at 30000 g for 30 min at 4°C. The supernatant was the osmotic shock fluid containing periplasmic proteins and was divided into two equal parts. One part was dialyzed extensively against Protein L Binding Buffer (100 NaH2PO4, pH 7.2; 150 mM NaCl), and the second part against IMAC Binding Buffer (50 mM NaH2PO4 pH 8.0, 500 mM NaCl, 15% glycerol, and 0.05% Tween 20). The dialysis was carried out at 4°C overnight before continuing with the purification. The supernatant was centrifuged after dialysis (30000 g, 30 min, 4°C).
Ni-NTA Agarose: The clear supernatant was collected and applied to immobilized-metal affinity chromatography (IMAC) with a 5 ml Ni-NTA Agarose (Qiagen) column equilibrated with IMAC Binding Buffer. The column was then washed sequentially with IMAC Washing Buffer (50 mM NaH2PO4 pH 8.0, 500 mM NaCl, 15% glycerol, 0.05% Tween 20, and 20 mM imidazole) and IMAC Elution Buffer (50 mM NaH2PO4 pH 8.0, 500 mM NaCl, 15% glycerol, 0.05% Tween 20, and 300 mM imidazole). The flow rate during application, washing and elution was 1.0 ml/min. The purification of scFv was examined by running an aliquot of the collected samples on SDS-PAGE and then staining with Coomassie Brilliant Blue. The purified proteins were pooled and dialyzed against 1xPBS at 4°C overnight. The proteins were kept at -80°C after determination of the concentration by a Bradford assay.
Protein L Agarose: The extract of E.coli scFvs was loaded onto a column filled with 5 ml of protein L agarose (Thermo Fisher Scientific) equilibrated with Protein L Binding Buffer at a flow rate of 1.0 ml/min. After washing with 10 column volumes of binding buffer, the proteins were eluted from the column with elution buffer that contained 0.1 M glycine at pH 2.5 (at a flow rate of 1.0 ml/min). Immediately, the eluted fractions were adjusted to physiological pH by adding 100 μL of neutralization buffer (1 M Tris, pH 8.0) to 1 ml of eluate. Fractions containing scFvs (as judged by SDS-PAGE and Coomassie Brilliant Blue staining) were pooled and dialyzed against 1xPBS at 4°C overnight. The concentration of protein was calculated by a Bradford assay. The purified fractions were stored at -80°C.
Protino Ni-IDA 1000: A clear mixture of proteins from the periplasm of E.coli was dialyzed against the 1x LEW buffer (Lysis-Equlibration-Wash Buffer, 50 mM NaH2PO4 pH 8.0, 300 mM NaCl.) Onto the equilibrated column (with 1xLEW buffer) filled with Ni-IDA 1000 (Macherey-Nagel), a fraction of periplasmic protein was applied (at flow rate 1.0 ml/min). Subsequently, the weakly bound proteins were washed away (1xLEW buffer, 5 x column volume, flow rate 1.5 ml/min), and elution was performed using a mixture of 1xLEW + 10 mM imidazole buffer and 1xLEW + 250 mM imidazole buffer with an imidazole gradient (up to 250 mM imidazole) (at a flow rate of 1.5 ml/min). From the fractions obtained after elution, samples for SDS-PAGE were prepared. The protein concentration was determined using the Bradford method. The fractions in which the presence of protein was found were dialyzed against 1xPBS and frozen at -80°C.
Protein A Antibody Purification: A sample containing proteins from the periplasmic fraction was diluted 2 x with binding buffer (included in the purification kit). The purification procedure was carried out according to the protocol provided by the manufacturer (Sigma-Aldrich).
Insect cells expression system: High Five cells were lysed by lysis buffer (50 mM Tris pH 7.5, 10% glycerol, 1% Triton X-100, 150 mM NaCl, 10 mM MgCl2, and 1 x Protease Inhibitor Cocktail) and shaken for 10 min in an ice bath. After lysis, the cell suspension was centrifuged (30000 g, 30 min, 4°C). The purification procedures for scFvs from insect cells with Ni-NTA Agarose, Protino Ni-IDA 1000 and Protein L Agarose were carried out in the same manner as previously described for bacterial scFvs.
Purification of insect antibodies with the c-myc tag: The cultures of High Five or Sf9 insect cell clones were thawed on ice. The pellet was suspended in lysis buffer (50 mM NaH2PO4, 500 mM NaCl, 15% glycerol, and 0.05% Tween 20 at pH 8.0 with the addition of 1 x protease inhibitors) and incubated on ice for 15 min. The sample was then centrifuged (30000 g, 10 min, 4°C), and the supernatant was transferred to a spin-column containing Pierce™ Anti-c-Myc Agarose and incubated for 2.5 hours with gentle stirring in a cold room. After incubation, the sample was centrifuged (14000 g, 10 s, RT). The spin columns were rinsed 3x times with TBS-T buffer (25 mM Tris, 0.15 M NaCl, pH 7.2 + 0.05% Tween) and 1x conditioning buffer (Thermo Fisher Scientific). The elution was carried out using two methods: (a). with Elution Buffer pH 2 (Thermo Fisher Scientific) and centrifugation (14 000 g, 10 s, RT), repeated 3 times in total, followed by addition of 1 M Tris pH 9.5 to the sample with elution buffer; and (b). By addition to a spin column with 25 μL Lane Marker Non-Reducing Sample Buffer 2x (prepared from 5x concentrate, Thermo Fisher Scientific), mixing and incubation for 5 min at 99°C, followed by centrifugation (14 000 g, 10 s, RT).
SDS-PAGE and Western blotting: Samples for analysis were mixed with 2x electrophoresis buffer containing 50 mM Tris-HCl at pH 6.8, 10% glycerol, 2% SDS, 0.1% bromophenol blue and 5% β-mercaptoethanol and heated for 5 min at 95°C. SDS-PAGE was performed according to Laemmli (1970) [18], with a 4% stacking gel and a 12% resolving gel (Mini-Protean 3, Bio-Rad). Gels were stained by Coomassie Brilliant Blue.
In the immunodetection identification experiment, protein bands from the SDS-PAGE gel were transferred to a PVDF membrane immediately after separation. Electrotransfer was performed in ice cold buffer (25 mM Tris-HCl at pH 8.3, 192 mM glycine, and 20% (v:v) methanol) at 100 V for 1 h. The PVDF membrane was then blocked in TBST buffer (20 mM Tris, pH 7.5, 150 mM NaCl, and 0.1% Tween 20) with 5% w/v non-fat dry milk for 1 h at room temperature. Afterward, the membrane was washed with TBST buffer three times for 10 min and incubated with Anti-c-Myc primary antibody (Sigma C3956) in blocking buffer overnight at 4°C on a shaker. The membrane was washed with TBST three times for 10 min at room temperature, and secondary antibody (Santa Cruz, sc-2030) in blocking buffer was added for 1 h. Then, the membrane was washed with TBST three times for 10 min. Finally, the membrane was incubated with Santa Cruz Biotechnology Luminol reagent for 1 min and visualized using Fusion FX5/quantum system (Vilber Lourmat).
Analysis of the specificity of the obtained scFvs monoclonal antibodies
The specificity of the purified scFvs was determined by ELISA. One hundred ml aliquots of purified antibodies at different concentrations resuspended in 4% BSA were incubated for 1.5 h at 37°C in 96-V-well plate. Then, 1.5 x105 CHO+ cells (expressing D2-5-HT1A heteromers) as well as CHO- cells (expressing D2R, 5-HT1AR) or CHO-K1 cells (resuspended in 50 ml of RPMI with 5% FBS) (Sigma-Aldrich) were added to the appropriate wells and incubated on ice for 1 h. The washing step was conducted 3 times at 4°C using 200 ml cold PBS, and after each round, the cells were centrifuged (300 g, 10 min, 4°C) and the supernatant was aspirated off. Then, the pellets were resuspended in 50 ml protein L conjugated with HRP (protein L was resuspended in 0.5% BSA/PBS at a 1:2000 ratio) (GE Healthcare) and incubated for 30 min on ice. After washing (carried out 4 times as described above), the reactions were developed by TMB substrate (GE Healthcare). Experiments were performed in triplicate.
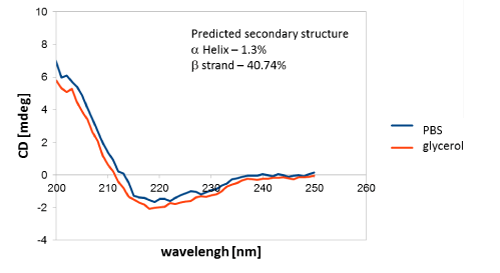
Circular dichroism measurements (CD Measurements)
Circular dichroism (CD) spectra were measured with a JASCO 710 spectropolarimeter at 20°C in a 0.1 cm cuvette. The spectra were run from 190 to 250 nm. Each spectrum was recorded at least three times and averaged. The final concentrations of the antibodies were 0.2 mg/mL in 1xPBS and protein stabilizing cocktail, which helps extend the shelf-life of most proteins. CD measurements were aimed at determining the content of secondary structures in the tested antibodies and investigating their stability.
Results and Discussion
Phage display technology is a very useful tool for the isolation of target-specific monoclonal antibodies. Unfortunately, the relatively fast selection process does not always coincide with the fast expression and purification of the protein. There are a number of different strategies (such as bacterial cells [19,20], mammalian cells [21], yeast [22], plant cells [23], and insect cells [24]) used to express scFv antibody fragments. The proteins can be expressed as correctly folded and directly active proteins or as aggregates requiring in vitro refolding to become active [16]. There are some general regulations to consider in the design of vectors and expression systems used with the different hosts, and each of the hosts has advantages and disadvantages for the production of active scFv antibodies [17,25]. Additionally, a wide range of methodologies has been adapted for successful purification of the given protein [17]. Nevertheless, success depends to a large extent on the properties of the antibody itself, which we are not able to predict in the early stages of selection. A problem that often accompanies the preparation of antibodies is their instability. Because of the nature of scFv fragments, this kind of antibody is less stable than the whole antibody. For example, immunoglobulin G (IgG) is a heterotetrameric molecule consisting of two heavy and two light chains connected via disulfide bonds. Heavy and light chains (HC and LC) also contain intramolecular disulfide bonds for stabilization [26]. In comparison, scFv consists only of variable (V) regions, and it possesses a soluble and flexible amino acid peptide linker which connects the V regions [7] and one disulfide bond for stabilization.
Because of the problems mentioned above, in the present work, we focused on the comparison of some strategies used to produce and purify soluble scFv antibodies. Two different expression systems - bacterial and insect cells - were adopted for this purpose. In both cases, (E.coli and insect cells), scFvs antibodies were successfully obtained (Figure 1).
There are three locations from which bacterially expressed proteins can be isolated: the cytoplasm, the periplasm, and the culture supernatant. Many studies showed that the majority of proteins are expressed as insoluble inclusion bodies in the cytoplasm of E.coli [27,28]. The purification of inclusion bodies is often interconnected with denaturation and refolding, which is problematic as far as the production of active soluble scFv antibodies is concerned [16]. In this study, soluble scFv antibodies were expressed in non-suppressive E. coli HB2151 bacteria cells. There is a termination codon between the phage pIII gene and the scFv gene. When the phage infects non-suppressive E. coli HB2151 cells, the termination codon will be translated. Therefore, the scFv gene will not be expressed as a fusion protein with pIII in HB2151 cells, but it will be expressed as the soluble protein and secreted into the periplasmic zone. In the E. coli expression system, the periplasmic compartment is preferably used for recombinant protein production. There appears to be much less protease activity in the periplasmic space than in the cytoplasm. In addition, scFv antibodies purification is simpler due to fewer contaminating proteins in the periplasm. Another advantage is that the correct formation of disulfide bonds can be facilitated in this location because the periplasmic space provides a more oxidative environment than does the cytoplasm, and it is rich in proteins which are important for folding and catalysing disulfide bond formation (PDI, DsbA and DsbC) or chaperones [29,30].
On the other hand, the use of a baculovirus expression system is a promising way to produce molecules for therapeutic purposes because of its capability of posttranslational modifications and absence of molecules toxic to humans, since the baculoviruses have a highly restricted host range. This was the first system that linked the advantages of bacterial systems, such as high production levels and scalability, with protein processing typically used in eukaryotic cells. Furthermore, insect cells can be grown on serum-free media, which is an advantage in terms of biosafety as well as cost. Increasing understanding of insect cell biology has led to technological advances in baculovirus production strategies over the last 30 years. One of the milestones that led to simplification and indefectibility was the deletion of an essential gene and insertion of the bacterial origin of replication into the viral genome, along with subsequent development of the E. coli DH10Bac strain containing the baculovirus genome as a part of the bacmid and a helper plasmid carrying a transposase gene. Transfection of these cells with transfer plasmids containing genes of interest controlled by polyhedrin promoter, flanked by sequences recognized by the transposase, causes transposition of the gene of interest into the polyhedrin locus of the bacmid, which contains matching Tn7 sequences. As a result of this process, the lacZ gene is deleted from the bacmid DNA, which enables one to use blue-white screening to isolate the recombinant bacmid DNA [31]. This kind of in vivo transposition guaranties 100% efficiency in producing recombinant baculovirus DNA, and this approach, known as the Bac-to-Bas system, was used in this study. A baculovirus carrying cDNA of scFV antibody was created and amplified in Sf9 cells. The high-titer baculovirus was used to infect High Five cells, which are considered to be more efficient at producing recombinant antibodies than Sf9 cells [32]. In preliminary experiments, both the multiplicity of infection and the time of cell growth after infection were tested. These optimization steps showed that satisfactory scFv production was achieved when the MOI (multiplicity of infection) amounted to 5 and cells were grown for 48 hours after infection. In conclusion, this stage of the studies showed that both investigated expression systems are suitable for scFv production.
In the next step, we compared a few methods of the purification of scFvs expressed as soluble proteins in E. coli and insect cells using affinity chromatography. Application of different methodologies available for antibody purification is widespread, as underlined in some thematic papers, e.g. [33–35]. Protein L agarose purification is a one-step purification method that yields significant amounts of pure protein. Protein L is an immunoglobulin-binding protein that was isolated from the bacteria Peptostreptococcus magnus and is now produced as a recombinant protein. Protein L binds to scFv without interfering with the antigen binding site. However, problems may occur during protein elution because the process requires very low-pH buffer (in our case, pH 2), which may have an adverse impact on protein stability. Protein A is a cell wall component produced by several strains of Staphylococcus aureus. The 46.7-kDa protein consists of a single polypeptide chain that is essentially devoid of carbohydrates. Native protein A contains four high-affinity binding sites (Ka = 108/mol) that are capable of interacting with the Fc region of IgG-class antibodies from selected mammalian species. The IgG-binding function is optimal at pH 8.2, but efficient binding also occurs in neutral and physiological buffers (pH 7.0 to 7.6). The IMAC with Ni-NTA Agarose and Protino Ni-IDA 1000 requires the presence of a His tag for efficient protein purification. Based on theoretical assumptions, all techniques mentioned above should yield positive effects, and pure, soluble protein should be obtained. Unfortunately, despite good validation, not all techniques gave satisfactory results. A fraction containing purified scFvs was not obtained with the immobilized-A protein beads. Similar results were obtained in the case of IMAC methodology—the purified antibody was obtained at very low concentrations. Only the purification of scFv antibodies by affinity chromatography with the L protein gave positive results, and highly concentrated protein samples were obtained (Figure 2).
Additionally, the purification results were confirmed by Western blot analysis (Figure 3).
Moreover, following scFv expression in insect cells, attempts to purify antibodies with an attached c-myc tag were undertaken, but unfortunately, yielded no success. Therefore, the results indicate that the purification of scFv antibodies only by affinity chromatography with L protein gave satisfactory results.
Additionally, CD measurements confirmed the predominant content of secondary structures in the tested antibodies (Figure 4), which was consistent with the literature data [36].
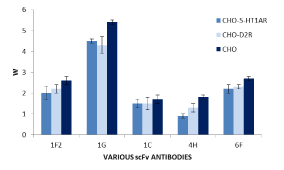
The analysis of specificity of the obtained scFvs monoclonal antibodies showed positive results (Figure 5). The antibodies with increased affinity to the given heteromer were obtained.
The major problem that occurred during the experiments was connected with the maintenance of protein stability, especially during prolonged storage. To overcome this problem, protein stabilizing cocktail was added to the purified antibodies. It has been widely accepted that solutions of enzymes and other proteins to which the cocktail is added may be refrigerated or frozen for storage without losing their activity or function. Unfortunately, this method did not work satisfactorily in our case.
Conclusion
The speed, along with the in vitro nature of the selection process and the ability to produce human antibodies, makes phage display a very potent technology for the generation of scFvs antibodies suitable for therapeutic development [5]. However, despite the numerous benefits of the methods, problems associated with obtaining soluble and active forms of proteins often arise. Therefore, in the present work, we compared prokaryotic and eukaryotic expression systems for the production of soluble scFv antibodies. Both systems are useful tools. In both the bacterial and insect cell expression systems, protein L agarose chromatography gave satisfactory results, and in our opinion, it is the best choice as a purification resin.
The manuscript has been corrected by AJE - Certificate Verification Key 414B-1A48-2CC6-B7D2-09D2. The Faculty of Biochemistry, Biophysics and Biotechnology is partner with the Leading National Research Centre (KNOW) supported by the Ministry of Science and Higher Education. This work was supported by grants 35+ Nr. 35p/1/2016 from KNOW.
- Bird RE, Hardman KD, Jacobson JW, Johnson S, Kaufman BM, et al. (1998) Single- chain antigen-binding proteins. Science 242: 423–426. Link: https://tinyurl.com/y6tphe7f
- Schwimmer LJ, Huang B, Giang H, Cotter RL, Chemla-Vogel DS, et al. (2013) Discovery of diverse and functional antibodies from large human repertoire antibody libraries. J Immunol Methods 391: 60-71. Link: https://tinyurl.com/yy27p2ln
- Chan CE, Chan AH, Lim AP, Hanson BJ (2011) Comparison of the efficiency of antibody selection from semi-synthetic scFv and non-immune Fab phage display libraries against protein targets for rapid development of diagnostic immunoassays. J Immunol Methods 373: 79-88. Link: https://tinyurl.com/y6os97qd
- Hoen PA, Jirka SM, Ten Broeke BR, Schultes EA, Aguilera B, et al. (2012) Phage display screening without repetitious selection rounds. Anal Biochem 421: 622-631. Link: https://tinyurl.com/y3f7pufz
- Shim H (2017) Antibody Phage Display, Springer International Publishing AG. Springer Nature.
- Tanabe A, Nakano K, Nakakido M, Nagatoishi S, Tanaka Y, et al. (2018) Production and characterization of a novel site-specific-modifiable anti-OX40-receptor single-chain variable fragment for targeted drug delivery. Biochem Biophys Res Commun 496: 614-620. Link: https://tinyurl.com/y4gvopsh
- Frenzel A, Hust M, Schirrmann T (2013) Expression of recombinant antibodies. Front Immunol 4: 217. Link: https://tinyurl.com/y2gazcad
- Barbas CF (2001) Phage display: a laboratory manual, Cold Spring Harbor, NY. Cold Spring Harbor Laboratory Press. Link: https://tinyurl.com/yyx9gaev
- Lee CM, Iorno N, SierroF, Christ D (2007) Selection of human antibody fragments by phage display. Nat Protoc 2: 3001–3008. Link: https://tinyurl.com/y57yn2et
- Smith GP (1985) Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228: 1315-1317. Link: https://tinyurl.com/y6sp6tqp
- Hoogenboom HR (2005) Selecting and screening recombinant antibody libraries, Nat Biotechnol 23: 1105-1116. Link: https://tinyurl.com/y5t2ghzq
- Miller KD, Weaver-Feldhaus J, Gray SA, Siegel RW, Feldhaus MJ (2005) Production, purification, and characterization of human scFv antibodies expressed in Saccharomyces cerevisiae, Pichia pastoris, and Escherichia coli. Protein Expr Purif 42: 255-267. Link: https://tinyurl.com/y3x58s6v
- Jefferis R (2009) Recombinant antibody therapeutics: the impact of glycosylation on mechanisms of action. Trends Pharmacol Sci 30: 356-362. Link: https://tinyurl.com/y4avxwlf
- Sakamoto S, Taura F, Tsuchihashi R, Putalun W, Kinjo J, et al. (2010) Expression, purification, and characterization of anti-plumbagin single-chain variable fragment antibody in Sf9 insect cell. Hybridoma 29: 481-488. Link: https://tinyurl.com/y32at9fd
- Wang Y, Zhang X, Zhang C, Liu Y, Liu X (2012) Isolation of single chain variable fragment (scFv) specific for Cry1C toxin from human single. Toxicon 60: 1290-1297. Link: https://tinyurl.com/y6kdq826
- Ahmad ZA, Yeap SK, Ali AM, Ho WY, Alitheen NB, et al. (2012) scFv antibody: principles and clinical application. Clin Dev Immunol 2012: 980250. Link: https://tinyurl.com/y2m7hgzv
- Verma R, Boleti E, George AJ (1998) Antibody engineering: comparison of bacterial, yeast, insect and mammalian expression systems. J Immunol Methods 216: 165-181. Link: https://tinyurl.com/y6bxen9z
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. Link: https://tinyurl.com/yxwelzxs
- Skerra A, Plückthun A (1988) Assembly of a functional immunoglobulin Fv fragment in Escherichia coli. Science 240: 1038–1041. Link: https://tinyurl.com/yyah8npt
- Mergulhão FJM, Monteiro GA, Cabral JMS, Taipa MA (2004) Design of bacterial vector systems for the production of recombinant proteins in Escherichia coli. J Micro and Biotech 14: 1–14. Link: https://tinyurl.com/y39gdq2c
- Ho M, Nagata S, Pastan I (2006) Isolation of anti-CD22 Fv with high affinity by Fv display on human cells. PNAS 103: 9637–9642. Link: https://tinyurl.com/yyh86ejg
- Wang Y, Shan Y, Gao X, Gong R, Zheng J, et al. (2018) Screening and expressing HIV-1 specific antibody fragments in Saccharomyces cerevisiae. Mol Immunol 103 279-285. Link: https://tinyurl.com/yxn52xg3
- Galeffi P, Lombardi A, Pietraforte I, Novelli F, Di Donato M, et al. (2006) Giacomini, Functional expression of a single-chain antibody to ErbB-2 in plants and cell-free systems. J Transl Med 4: 39. Link: https://tinyurl.com/y64gqn7v
- Choo ABH, Dunn RD, Broady KW, Raison RL (2002) Soluble expression of a functional recombinan cytolytic immunotoxin in insect cells. Prot Expres and Purific 24: 338–347. Link: https://tinyurl.com/y2dwdubc
- Knaust RKC, Nordlund P (2001) Screening for soluble expression of recombinant proteins in a 96-well format. Analyt Biochem 297: 79–85. Link: https://tinyurl.com/y43b5xqw
- Edelman GM (1973) Antibody structure and molecular immunology. Science 180: 830–840. Link: https://tinyurl.com/y6nxb7mc
- Sushma K, Vijayalakshmi MA, Krishnan V, Satheeshkumar PK (2010) Cloning, expression, purification and characterization of a single chain variable fragment specific to tumor necrosis factor alpha in Escherichia coli. J Biotechnol 156: 238-244. Link: https://tinyurl.com/yxkoycss
- Zhu X, Wang D, Li S, Xiao Q, Tao K, et al. (2012) Expression of a humanized single-chain variable fragment antibody targeting chronic myeloid leukemia cells in Escherichia coli and its characterization. Int J Mol Med 29 939-945. Link: https://tinyurl.com/y2n2ym6p
- Choi JH, Lee SY (2004) Secretory and extracellular production of recombinant proteins using Escherichia coli. Appl Microbiol Biotechnol 64: 625-635. Link: https://tinyurl.com/y4bd6x87
- Dewi KS, Retnoningrum DS, Riani C, Fuad AM (2016) Construction and Periplasmic Expression of the Anti-EGFRvIII ScFv Antibody Gene in Escherichia coli. Sci Pharm 84: 141-152. Link: https://tinyurl.com/y5xvmxvo
- Luckow VA, Lee SC, Barry GF, Olins PO (1993) Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J Virol 67: 4566-4579. Link: https://tinyurl.com/y5qxckg2
- Furuta T, Ogawa T, Katsuda T, Fujii I, Yamaji H (2010) Efficient production of an antibody Fab fragment using the baculovirus-insect cell system. J Biosci Bioeng 110: 577-581. Link: https://tinyurl.com/y567s48n
- Link: https://tinyurl.com/h73yztz
- Rodrigo G, Gruvegård M, Van Alstine JM (2015) Antibody Fragments and Their Purification by Protein L Affinity Chromatography. Antibodies 4: 259-277. Link: https://tinyurl.com/y2o8d3bk
- Darcy E, Leonard P, Fitzgerald J, Danaher M, MaH, et al. (2017) Purification of Antibodies Using Affinity Chromatography. Methods Mol Biol 1485: 305-318. Link: https://tinyurl.com/y37jopw3
- Conroy P, Law R, Gilgunn S, Hearty S, Caradoc-Davies T, et al. (2014) Reconciling the structural attributes of avian antibodies. J Biol Chem 289: 15384-15392. Link: https://tinyurl.com/y3mq2qp9
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.




 Save to Mendeley
Save to Mendeley
