International Journal of Veterinary Science and Research
Pacific bioscience sequence technology: Review
Abde Aliy Mohammed*, Bayeta Senbeta and Takale Worku
Cite this as
Mohammed AA, Senbeta B, Worku T (2022) Pacific bioscience sequence technology: Review. Int J Vet Sci Res 8(1): 027-033. DOI: 10.17352/ijvsr.000108Copyright License
© 2022 Mohammed AA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Pacific Biosciences has developed a platform that may sequence one molecule of DNA in a period via the polymerization of that strand with one enzyme. Single-molecule real-time sequencing by Pacific BioSciences’ technology is one of the most widely utilized third-generation sequencing technologies. PacBio single-molecule real-time Sequencing uses the Zero-mode waveguide’s ingenuity to distinguish the best fluorescence signal from the stable fluorescent backgrounds generated by disorganized free-floating nucleotides. PacBio single-molecule real-time sequencing does not require PCR amplification, and the browse length is a hundred times longer than next-generation sequencing. It will only cover high-GC and high-repeat sections and is more accurate in quantifying low-frequency mutations. PacBio single-molecule real-time sequencing will have a relatively high error rate of 10%-15% (which is practically a standard flaw of existing single-molecule sequencing technology). In contrast to next-generation sequencing, however, the errors are unintentionally random. As a result, multiple sequencing will effectively rectify the bottom deviance. Unlike second-generation sequencing, PacBio sequencing may be a technique for period sequencing and doesn’t need an intermission between browse steps. These options distinguish PacBio sequencing from second-generation sequencing, therefore it’s classified because of the third-generation sequencing. PacBio sequencing produces extremely lengthy reads with a high error rate and low yield. Short reads refine alignments/assemblies/detections to single-nucleotide precision, whereas PacBio long reads provide reliable alignments, scaffolds, and approximate detections of genomic variations. Through extraordinarily long sequencing reads (average >10,000 bp) and high accord precision, the PacBio Sequencing System can provide a terribly high depth of genetic information. To measure and promote the event of modern bioinformatics tools for PacBio sequencing information analysis, a good browse machine is required.
Introduction
By providing significantly longer reads, single-molecule sequencing reduced composition bias, and a slip-up profile that is orthogonal to alternative platforms, Pacific Biosciences technology has the potential to overcome some of the shortcomings of current next-generation sequencing platforms [1]. One of the most widely utilized third-generation sequencing technologies is PacBio’s SMRT (single molecule real-time) sequencing [2].
Although Pacific Biosciences (PacBio) is a less expensive platform (per run) and produces much longer reads (3,000 to 15,000 bp without requiring a library preparation amplification step), a recent review found that PacBio was, in theory, the least appropriate of the major high-throughput sequencing platforms for biological process identification [1], owing to its low accuracy. Inferiority reads are difficult since biological process identification requires high read accuracy; however, this issue can be alleviated by using PacBio circular consensus sequencing. While Second-Generation Sequencing (SGS) technologies have provided significant improvements over Sanger sequencing, their limitations, particularly their short read lengths, make them unsuitable for some specific biological concerns, as well as the assembly and identification of complex genomic areas, the detection of sequence isoforms, and the detection of methylation. Pacific Biosciences (PacBio) developed single-molecule period (SMRT) sequencing as an alternative to overcome some of these restrictions [2].
To put it succinctly, it’s known as “PacBio sequencing”; nevertheless, the community also refers to it as “SMRT sequencing.” PacBio sequencing, unlike SGS, may be a period sequencing technology that does not require a pause between reading steps. PacBio sequencing is distinguished from SGS by these features, hence it is classified as Third-Generation Sequencing (TGS). PacBio sequencing allows for significantly longer read lengths and faster runs than SGS methods, but it is limited by lesser yield, a higher error rate, and a lower cost per base. Because the advantages of PacBio sequencing and SGS are mutually beneficial. The complementary strengths and weaknesses of SGS and PacBio sequencing prompted a unique concept, hybrid sequencing, to blend the two techniques [1].
These methods often entail using high-throughput, high-accuracy short browse information to correct errors in long reads, in order to reduce the amount of expensive long-read sequence knowledge required and to save the comparably long, but more fallible, sub reads. Furthermore, PacBio long reads provide reliable alignments, scaffolds, and preliminary detections of genomic variations, whereas short reads refine alignments/assemblies/detections to single-nucleotide precision [3,4].
SGS knowledge’s broad scope can be used in downstream mensuration. In general, PacBio sequencing produces extremely long reads with a high error rate and low yield. Its relative performance in comparison to first-generation, second-generation, and third-generation sequencing technologies. PacBio RS II, which uses the sixth generation of enzymes and the fourth generation of chemistry (P6-C4), has a longer average browse time than SGS platforms, but a lower yield and a lower single-pass mistake rate. Furthermore, PacBio sequencing is both faster and more expensive than most other methods [4]. Internal control options in bioinformatics workflows used to preprocess raw sequences before biological process analysis are aimed to reduce sequencing or PCR errors in the dataset. Reads with an uncertain base decision, a mean quality score below a threshold, multiple mismatches to a primer/barcode sequence, less than a certain number of bases, or chimeras, for example, are removed from processes [5]. With categorization binning, the PacBio sequencing platform was suitable for biological process identification of electrically generated microbiomes down to the genus level [6]. Circular consensus Sequences were used to overcome the low browsing quality that is typical of PacBio. In addition, internal control workflows were modified to address PacBio-specific issues, the most significant of which was the creation of ‘PacBio chimeras,’ alternatives that may be the target of political action committees. Chime, on the other hand, does not appear to detect bio library preparation. Future advances by political action committees will be similar to those made by each sequencing platform. Biotechnology and chemistry can change lengthier (thus a lot of correct and numerous) reads, whilst understanding of political action committees can change shorter (hence a lot of accurate and numerous) reads. Bio biases can skew a lot of correct knowledge when it comes to identifying biological processes [7].
Principles of pacbio sequencing technology
Examine hybrid-sequencing methods that combine first and second-generation sequencing technology to overcome the drawbacks of each separately, as well as the applicability of PacBio sequencing to a variety of areas of research, including ordering, transcriptome, and epigenetics. Due to new procedural approaches and advancements in sequencing technology, PacBio sequencing will now be used to analyze a wide range of bigger genomes, including human genomes [4].
Through very long sequencing reads (average >10,000 bp), the PacBio Sequencing System may provide a tremendous amount of genetic information, resulting in high consensus accuracy. Is the fluorescently tagged ester detected because it is integrated by the deoxyribonucleic acid enzyme into the complementary strand of the one molecule? The fluorescent label specific for the bottom is detected at the time of integration, while the enzyme simultaneously chops off the Label-from-the-Ester. The process is repeated for each consecutively labeled ester, and the bottom sequence is determined by the order of the four completely different labels identified.
The procedure is carried out in chambers with zero-mode waveguides (ZMW). A single molecule of the enzyme is immobilized and a fluorescently tagged ester is added at intervals between the ZMWs, allowing deoxyribonucleic acid sequencing to be evaluated optically and recorded in real-time [8]. The ZMW invention is employed by PacBio SMRT Sequencing to distinguish the ideal fluorescent signal from the powerful fluorescent backgrounds created by unstructured free-floating nucleotides. A deoxyribonucleic acid enzyme’s binding, and therefore the guide deoxyribonucleic acid strand, is anchored to the ZMW’s lowest glass surface. Because the ZMW dimensions are lower than the wavelength of the sunlight, the optical device lightweight goes past the lowest surface of a ZMW but does not entirely pierce it. As a result, it is possible to selectively excite and identify sunlight emitted by nucleotides recruited for base elongation [4,9,10] Figure 1.
DNA sequencing and pacbio library construction
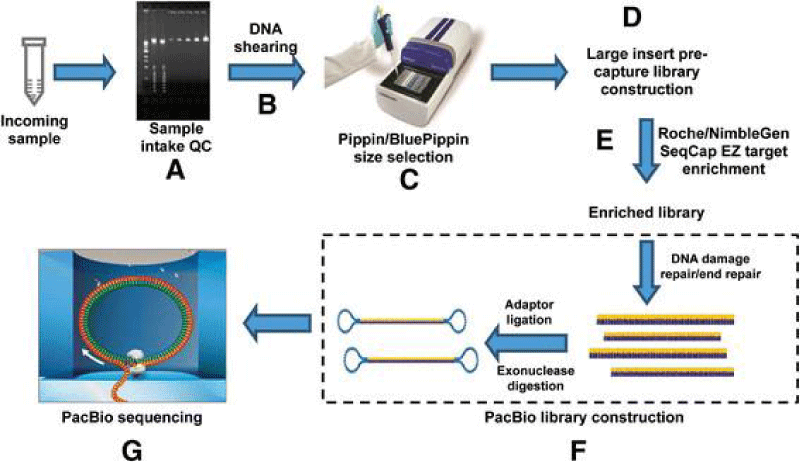
Genomic DNA was sheared with an ultrasonicator and reborn into the exclusive SMRT bell TM library format with the help of the RS DNA guide Preparation Kit. Sheared DNA was repaired completely, and pin adapters were ligated using T4 DNA ligase. Nuclease III and nuclease VII were used to destroy incompletely formed SMRT bell templates. The resulting DNA templates were refined using SPRI magnetic beads and subjected to a two-fold molar way over a sequencing primer that was unique to the pin adapters’ fiber loop region [11] Figure 2.
The following steps make up the library construction workflow:
- Determine the genomic deoxyribonucleic acid standard (gDNA)
- Using a g-TUBE to shear gDNA (Covaris)
- Choose a size and adjust the concentration
- Deoxyribonucleic acid damage and fragmented deoxyribonucleic acid ends are repaired.
- Purification of deoxyribonucleic acid
- Victimization of blunt adapters by blunt-end ligatures
- Cleanse the example before submitting it to a sequencer; the example, known as an SMRT bell, could be anything.
A circular deoxyribonucleic acid with a closed fiber generated by legating pin adapters to the ends of target double-stranded deoxyribonucleic acid (dsDNA) molecules [4,8]. Pacific Biosciences has created a technology that can sequence one molecule of deoxyribonucleic acid in a single period using a single Enzyme to polymerize that strand [12,13]. Individual picolitre-sized wells with clear bottoms are housed in specific flow cells in SMRT technology. Zero mode waveguides (ZMW) include one attached enzyme at an all-time low in each of the wells [13.14].
Because the enzyme integrates tagged bases onto the example deoxyribonucleic acid, one deoxyribonucleic acid molecule, which is circularized during library preparation (SMRT bell), can progress through the well. The incorporation of bases causes visible radiation to be emitted, which is recorded in the period through the ZMW’s glass bottoms [13-15]. At the start, the average browse length for SMRT was only ninety-one 5 Kb, with a high error rate of -13 percent due to incorrect insertions [16].
However, errors are randomly distributed over the browses [12], and excellent agreement sequences are frequently achieved with sufficient read depths [15,18]. Furthermore, every ester position during a 1kb amplicon is frequently browsed a hundred and ten times victimization circular consensus sequence methodology for one molecule with a hundred and ten kilobytes browse length, making it unlikely that the same random mistake would occur in multiple reads [19,20].
Advantage of pacbio sequence technology
PacBio long-read sequencing provided by SMRT Sequencing technology does not require PCR amplification, hence the read duration is a hundred times longer than NGS [7] compared to the previous two generations. PacBio SMRT sequencing can be utilized for genomic First State Novo sequencing to generate high-quality ordering sequences, obtain entire transcriptome information, and detect several junction isoforms, numerous mutations in target regions, and epigenetic alterations, among other things [7,8].SMRT sequencing may be accustomed to verifying the repeat size and therefore the detection of the quantity of interrupting AGG units [21]. The main advantage of this approach is that the unambiguous separation of the 2 CGG repeats on the various X chromosomes of females thereby outperforming all different (PCR) approaches. Afterward, the knowledge generated by SMRT sequencing is employed clinically for improved genetic guidance of girls deliberation the danger of getting a toddler with FXS [21-23]. Another example of braving a bicycle-built-for-two repeat by SMRT sequencing is that the ATTCT repeats embedded in deoxyribonucleic acid nine of the Spinocerebellar ataxy kind ten sequences [24]. The complete length of an enlarged ATTCT repeat was sequenced for the first time using SMRT technology for the first time. The repeat was recreated using assembly, with each interruption acknowledged and new interruptions discovered [24]. Whole-genome sequencing, targeted sequencing, enhanced population analysis, polymer sequencing of targeted transcripts, and microbic epigenetics have all been done with the PacBio RS II [8-10]. The circular consensus methodology has been defined as a preparative methodology useful in sequencing polymer viruses to extremely high accuracy, allowing the detection of extremely rare variants and accurate measurement of low-frequency variants [25]. The Pacific Biosciences platform’s lengthy sequence reads have allowed researchers to examine problematic sections of the ordering, such as MHC category I region transcripts [26,27] and regions of segmental duplication [28]. The impact of the platform and its potential involvement in the first State eloping routine analysis of human genomes driven by de novo assembly rather than comparisons to a reference sequence have also been demonstrated in studies undertaken to come up with first State Novo assemblies [29,30].
Application of pacbio sequencing technology and analysis of bioinformatics
Bioinformatics analysis, such as de novo assemblies to identify genetic variations, reference ordering mapping, ordering annotation (pathogenic and status genes prediction, non-coding polymer prediction, CRISPRs prediction), cistron operates annotation (COG/ GO/ KEGG), SNP/InDel identification, and comparative genetic science analysis, biological process analysis, and divergence time estimation are all viable [9,10]. For SMRT knowledge analysis, more and more bioinformatics tools and algorithms are being developed, such as sequence alignment programs BLASR [31] and GraphMap [32], genome computer programs canu [33] and miniasm [34], and structural variant callers PBHoney [35] and Sniffles [36]. Furthermore, PacBio sequencing has been rapidly developed with multiple variants. It’s critical that these tools are benchmarked and evaluated using reads simulated by sequencing simulators targeting a specific version of PacBio technology [37]. For PacBio sequencing knowledge analysis, an efficient read machine is critical to gauging and promoting the event of recent bioinformatics tools [38].
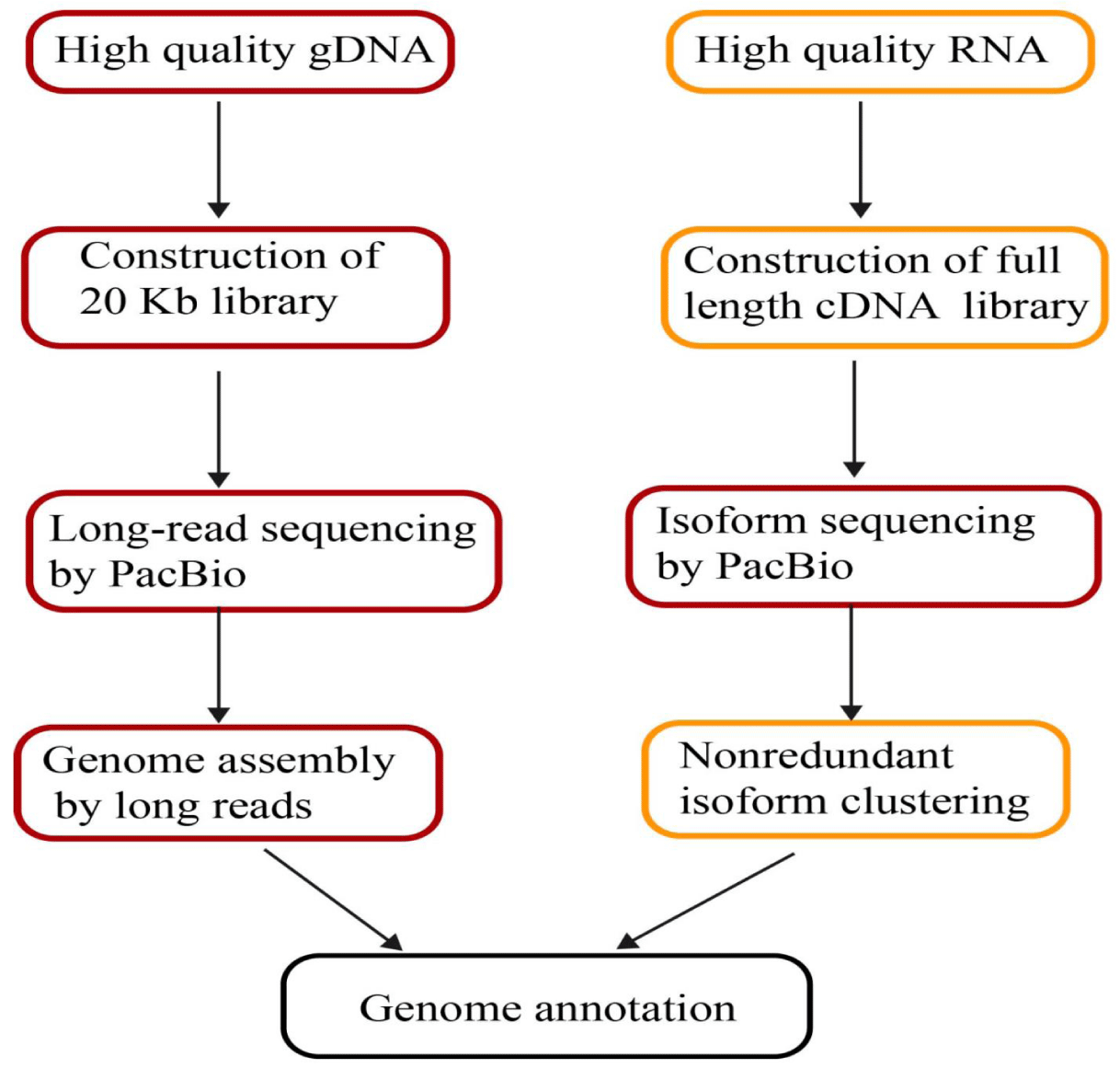
Single-molecule time sequencing could be used for a wide range of genetic research investigations. Reading lengths from the single-molecule period of time sequencing square measure corresponding to or larger than those from the Sanger sequencing technique backed dideoxynucleotide chain termination for de novo assemblies. The increased read length allows for Novo ordering sequencing and easier ordering assemblies in the United States [39,40]. To combine short-read sequence expertise with long-read sequence knowledge, scientists are using a single-molecule period of time sequencing in hybrid for de novo assemblies of genomes Table 1.
Many peer-reviewed publications demonstrating the automatic finishing of microorganism genomes were available for free in 2012, including one work that upgraded the Celera computer program with a pipeline for ordering finishing using lengthy SMRT sequencing reads [41]. Long-read sequencing was predicted in 2013 to be able to completely assemble and finish the majority of microbial and archaeal genomes [42]. By creating a circular polymer Template and using a strand displacing accelerator to detach the recently synthesized polymer strand from the template, the same polymer molecule can be re-sequenced several times [43].
The Broad Institute published an Associate in the Nursing examination of SMRT sequencing for SNP business in August 2012 [44]. The kinetics of an enzyme will reveal whether a base is an alkyl group or not [45]. Scientists have proven that single-molecule time sequencing may be used to investigate methylation and other base alterations [46-48]. SMRT sequencing was utilized by a group of scientists in 2012 to obtain the entire methylomes of six bacteria. A report on genome-wide methylation of an E. coli disease strain was published in November 2012 [49].
Long reads allow for the sequencing of whole cistron isoforms as well as the 5’ and 3’ ends. Isoforms and splice variants can be captured with this type of sequencing [50]. When investigating families with probable parental ductless gland disease, SMRT sequencing has various applications in procreative medical genetic science study. To determine the parent-of-origin of mutations, long readings modify haplotype phasing in patients. Deep sequencing allows for the determination of gene frequencies in spermatozoan cells, as well as the estimation of the risk of future offspring being affected [51-60].
Conclusion
PacBio sequencing allows for significantly longer browse lengths and faster runs than SGS but is limited by the poorer turnout, a higher error rate, and a higher expense per base. Long browse lengths (for Diamond State Novo assemblies of novel genomes), direct measuring of individual molecules, templates ready without PCR amplification, the system records the dynamics of every ester incorporation reaction, alter and improve genomic assembly and understanding of illness heritability are some of the benefits of the SMRT sequencing platform when compared to other sequencing technologies. Unwanted ground noise caused by biological building materials “is solved for the first time by zero-mode waveguiding technology.” With categorization binning, the PacBio sequencing platform proved sufficient for organic process identification of electro-generated microbiomes to the genus level. The rapid sequencing has also resulted in several evident flaws. For example, PacBio SMRT sequencing’s rather high error rate (which is practically a standard flaw of current single-molecule sequencing technology) will approach 10% -15%. In contrast to next-generation sequencing, however, the errors are unintentionally random. As a result, the bottom deviation is effectively corrected through multiple sequencing, and PacBio SMRT sequencing consensus accuracy is greater than 99.9%. (Q50). Once the ordination is sequenced, the base modification is directly identified. CD Genetic Science will provide integrated PacBio SMRT sequencing services, as well as long-read metagenomic sequencing and microorganism whole-ordination Novo sequencing. PacBio SMRT sequencing does not require PCR amplification, can cover high-GC and high-repeat regions, and is extremely accurate in quantifying low-frequency mutations. A good sequence machine can generate benchmark datasets with known ground truth, which can be used to assess the latest bioinformatics tools.
- Loman NJ, Constantinidou C, Chan JZ, Halachev M, Sergeant M, et al. (2012) High-throughput bacterial genome sequencing: an embarrassment of choice, a world of opportunity. Nat Rev Microbiol 10: 599-606.Link: https://bit.ly/3iHuyWb
- Koren S, Schatz MC, Walenz BP, Martin J, Howard JT, et al. (2012) Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol 30: 693-700. Link: https://bit.ly/3qFUEx2
- Schadt EE, Turner S, Kasarskis A (2010) A window into third-generation sequencing. Hum Mol Genet 19: R227-R240. Link: https://bit.ly/3uE92qK
- Rhoads A, Au KF (2015) Pacbio sequencing and its applications. Genomics Proteomics Bioinformatics 13: 278-289. Link: https://bit.ly/3qLlSSV
- Jumpstart Consortium Human Microbiome Project Data Generation Working Group (2012) Evaluation of 16S rDNA-based community profiling for human microbiome research. PLoS One 7: e39315. Link: https://bit.ly/3NrQnqN
- Marshall CW, Ross DE, Fichot EB, Norman RS, May HD (2012) Electrosynthesis of commodity chemicals by an autotrophic microbial community. Appl Environ Microbiol 78: 8412-8420. Link: https://bit.ly/3uu6BqM
- Fichot EB, Norman RS (2013) Microbial phylogenetic profiling with the Pacific Biosciences sequencing platform. Microbiome 1. Link: https://bit.ly/3NwufLS
- Ansorge WJ (2010) Next generation DNA sequencing techniques and applications. New Biotechnology 27: S3.
- Kong N, Ng W, Thao K, Agulto R, Weis A, et al. (2017) Automation of PacBio SMRTbell NGS library preparation for bacterial genome sequencing. Stand Genomic Sci 12. Link: https://bit.ly/36zRVyg
- Au KF, Underwood JG, Lee L, Wong WH (2012) Improving PacBio long read accuracy by short read alignment. PloS one 7: e46679. Link: https://bit.ly/35p9h0s
- English AC, Richards S, Han Y, Wang M, Vee V, et al. (2012) Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PloS one 7: e47768. Link: https://bit.ly/3qIQ0hM
- Eid J, Fehr A, Gray J, Luong K, Lyle J et al. (2009) Real-time DNA sequencing from single polymerase molecules. Science 323: 133-138. Link: https://bit.ly/3JSL6qe
- Ardui S, Ameur A, Vermeesch JR, Hestand MS (2018) Single molecule real-time (SMRT) sequencing comes of age: applications and utilities for medical diagnostics. Nucleic Acids Res 46: 2159-2168. Link: https://bit.ly/3uCVifP
- Levene MJ, Korlach J, Turner SW, Foquet M, Craighead HG, et al. (2003) Zero-mode waveguides for single-molecule analysis at high concentrations. science 299: 682-686. Link: https://bit.ly/36zNwLK
- Pollard MO, Gurdasani D, Mentzer AJ, Porter T, Sandhu MS (2018) Long reads: their purpose and place. Hum Mol Genet 27: R234-R241. Link: https://bit.ly/3NrXI9V
- Carneiro MO, Russ C, Ross MG, Gabriel SB, Nusbaum C, et al. (2012) Pacific biosciences sequencing technology for genotyping and variation discovery in human data. BMC genomics 13. Link: https://bit.ly/3tJHfGe
- Roberts RJ, Carneiro MO, Schatz MC (2013) The advantages of SMRT sequencing. Genome biology 14: 1-4. Link: https://bit.ly/36V8dSr
- Hebert PD, Braukmann TW, Prosser SW, Ratnasingham S, DeWaard JR, et al. (2018) A Sequel to Sanger: amplicon sequencing that scales. BMC genomics 19. Link: https://bit.ly/3JNDSn3
- Travers KJ, Chin CS, Rank DR, Eid JS, Turner SW (2010) A flexible and efficient template format for circular consensus sequencing and SNP detection. Nucleic Acids Res 38. Link: https://bit.ly/3DifYhk
- Hestand MS, Van Houdt J, Cristofoli F, Vermeesch JR (2016) Polymerase specific error rates and profiles identified by single molecule sequencing. Mutat Res 784: 39-45. Link: https://bit.ly/3IKcUeR
- Ardui S, Race V, Zablotskaya A, Hestand MS, Van Esch H, et al. (2017) Detecting AGG interruptions in male and female FMR1 premutation carriers by single‐molecule sequencing. Hum Mutat 38: 324-331. Link: https://bit.ly/3LrJqnZ
- Chen L, Hadd A, Sah S, Filipovic-Sadic S, Krosting J, et al. (2010). An information-rich CGG repeat primed PCR that detects the full range of fragile X expanded alleles and minimizes the need for southern blot analysis. J Mol Diagn 12: 589-600. Link: https://bit.ly/3uAy1vc
- Hayward BE, Usdin K (2017) Improved assays for AGG interruptions in fragile X premutation carriers. J Mol Diagn 19: 828-835. Link: https://bit.ly/3NtDgp2
- McFarland KN, Liu J, Landrian I, Godiska R, Shanker S, et al. (2015) SMRT sequencing of long tandem nucleotide repeats in SCA10 reveals unique insight of repeat expansion structure. PloS one 10: e0135906. Link: https://bit.ly/35kn4oZ
- Acevedo A, Andino R (2014) Library preparation for highly accurate population sequencing of RNA viruses. Nat Protoc 9: 1760-1769. Link: https://bit.ly/3IHz2Xm
- Chang CJ, Chen PL, Yang WS, Chao KM (2014) A fault-tolerant method for HLA typing with PacBio data. BMC Bioinformatics 15. Link: https://bit.ly/3uBMMOj
- Westbrook CJ, Karl JA, Wiseman RW, Mate S, Koroleva G, et al. (2015) No assembly required: Full-length MHC class I allele discovery by PacBio circular consensus sequencing. Hum Immunol 76: 891-896. Link: https://bit.ly/3iI7iqO
- Huddleston J, Ranade S, Malig M, Antonacci F, Chaisson M, et al. (2014) Reconstructing complex regions of genomes using long-read sequencing technology. Genome res 24: 688-696. Link: https://bit.ly/3K4WTlr
- Chaisson MJ, Wilson RK, Eichler EE (2015) Genetic variation and the de novo assembly of human genomes. Nat Rev Genet 16: 627-640. Link: https://bit.ly/3qKhm7k
- Erlich Y (2015) A vision for ubiquitous sequencing. Genome Res 25: 1411-1416. Link: https://bit.ly/3Dldqif
- Chaisson MJ, Tesler G (2012) Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics 13. Link: https://bit.ly/3ILYJWH
- Sović I, Šikić M, Wilm A, Fenlon SN, Chen S, et al. (2016) Fast and sensitive mapping of nanopore sequencing reads with GraphMap. Nat Commun 7. Link: https://bit.ly/35mMd2m
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, et al. (2017) Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome res 27: 722-736. Link: https://bit.ly/3tLNrgJ
- Li H (2016) Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32: 2103-2110. Link: https://bit.ly/3Dm2gtH
- English AC, Salerno WJ, Reid JG (2014) PBHoney: identifying genomic variants via long-read discordance and interrupted mapping. BMC Bioinformatics 15. Link: https://bit.ly/3INBVG6
- Sedlazeck FJ, Rescheneder P, Smolka M, Fang H, Nattestad M, et al. (2018) Accurate detection of complex structural variations using single-molecule sequencing. Nat Methods 15: 461-468. Link: https://bit.ly/36XzIe4
- Escalona M, Rocha S, Posada D (2016) A comparison of tools for the simulation of genomic next-generation sequencing data. Nat Rev Genet 17: 459-469. Link: https://bit.ly/3uEkrXA
- Zhao M, Liu D, Qu H (2017) Systematic review of next-generation sequencing simulators: computational tools, features and perspectives. Brief Funct Genomics 16: 121-128. Link: https://bit.ly/3JOJZYD
- Rasko DA, Webster DR, Sahl JW, Bashir A, Boisen N, et al. (2011) Origins of the E. coli strain causing an outbreak of hemolytic–uremic syndrome in Germany. N Engl J Med 365: 709-717. Link: https://bit.ly/3DmVUtX
- Chin CS, Sorenson J, Harris JB, Robins WP, Charles RC, et al. (2011) The Origin of the Haitian Cholera Outbreak Strain. N Engl J Med 364: 33–42. Link: https://bit.ly/3uFc2mC
- Koren S, Schatz MC, Walenz BP, Martin J, Howard JT, et al. (2012) Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol 30: 693–700. Link: https://bit.ly/3qGt9ne
- Koren S, Harhay GP, Smith TP, Bono JL, Harhay DM, et al. (2013) Reducing assembly complexity of microbial genomes with single-molecule sequencing. Genome Biol 14. Link: https://bit.ly/3IVba2I
- Smith CC, Wang Q, Chin CS, Salerno S, Damon LE, et al. (2012) Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature 485: 260-263. Link: https://bit.ly/36YNhdf
- Carneiro MO, Russ C, Ross MG, Gabriel SB, Nusbaum C, et al. (2012) Pacific biosciences sequencing technology for genotyping and variation discovery in human data. BMC genomics 13. Link: https://bit.ly/3uBzgu1
- Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, et al. (2010) Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods 7: 461-465. Link: https://bit.ly/3LlyDLV
- Clark TA, Murray IA, Morgan RD, Kislyuk AO, Spittle KE, et al. (2012) Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res 40: e29. Link: https://bit.ly/3tNne1n
- Song CX, Clark TA, Lu XY, Kislyuk A, Dai Q, et al. (2012) Sensitive and specific single-molecule sequencing of 5-hydroxymethylcytosine. Nature methods 9: 75-77.Link: https://go.nature.com/3IMS9zh
- Clark TA, Spittle KE, Turner SW, Korlach J (2011) Direct detection and sequencing of damaged DNA bases. Genome Integr 2. Link: https://bit.ly/3uzaZ7X
- Fang G, Munera D, Friedman DI, Mandlik A, Chao MC, et al. (2012) Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat Biotechnol 30: 1232-1239. Link: https://bit.ly/3IKo6rT
- Michael S, Fabian G, Hagen T, Donald S (2013) A single-molecule long-read survey of the human transcriptome. Nat Biotechnol 31: 1009– 1014. Link: https://bit.ly/3qIYzZY
- Ardui S, Ameur A, Vermeesch JR, Hestand MS (2018) Single molecule real-time (SMRT) sequencing comes of age: applications and utilities for medical diagnostics. Nucleic Acids Res 46: 2159-2168. Link: https://bit.ly/36vyIhr
- Wilbe M, Gudmundsson S, Johansson J, Ameur A, Stattin EL, et al. (2017) A novel approach using long‐read sequencing and ddPCR to investigate gonadal mosaicism and estimate recurrence risk in two families with developmental disorders. Prenat Diagn 37: 1146-1154. Link: https://bit.ly/3tL06AL
- Li C, Lin F, An D, Wang W, Huang R (2018) Genome sequencing and assembly by long reads in plants. Genes 9: 6. Link: https://bit.ly/3JO6yww
- Schadt EE, Turner S, Kasarskis A (2010) A window into third-generation sequencing. Hum Mol Genet 19: R227-R240. Link: https://bit.ly/3tPpyoU
- Korlach J, Bibillo A, Wegener J, Peluso P, Pham TT, et al. (2008) Long, processive enzymatic DNA synthesis using 100% dye-labeled terminal phosphate-linked nucleotides. Nucleosides Nucleotides Nucleic Acids 27: 1072-1083. Link: https://bit.ly/3tWBghz
- Korlach J, Bjornson KP, Chaudhuri BP, Cicero RL, Flusberg BA, et al. (2010) Real-time DNA sequencing from single polymerase molecules. Methods Enzymol 472: 431-455. Link: https://bit.ly/3uu7zDq
- McCarthy A (2010) Third generation DNA sequencing: pacific biosciences' single molecule real time technology. Chem Biol 17: 675-676. Link: https://bit.ly/3ISxXvQ
- Schadt EE, Turner S, Kasarskis A (2010) A window into third-generation sequencing. Hum Mol Genet 19: R227-R240. Link: https://bit.ly/3iLtPTS
- Levine MJ, Korlach J, Turner SW, Foquet M, Craighead HG, et al. (2003) Zero mode waveguides for single molecule analysis at high concentration. Science 299: 682-686. Link: https://bit.ly/3LrGurt
- Bashir A, Klammer AA, Robins WP, Chin CS, Webster D, et al. (2012) A hybrid approach for the automated finishing of bacterial genomes. Nat Biotechnol 30: 701-707. Link: https://bit.ly/3wM2HMP
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
