International Journal of Veterinary Science and Research
Research progress of gene editing technology CRISPR/Cas9 system in animal gene editing
Feng Fan1,2, Kang Mengna2, Zhu Xiaolan1, Cao Weiping1, Liu Yueqin1, Chen Qi1, Lin Fangfang1, Qianfu Yanwen1 and Xu Wenlina1*
2The Institute of Life Science, Jiangsu University, 301 Xuefu Road, Zhenjiang 212013, China
Cite this as
Fan F, Mengna K, Xiaolan Z, Weiping C, Wenlina X, et al. (2018) Research progress of gene editing technology CRISPR/Cas9 system in animal gene editing. Int J Vet Sci Res 4(1): 015-019. DOI: 10.17352/ijvsr.000030Gene editing technology, from the beginning of RNA interference (RNAi) technology to efficient developed enzyme technology, has been widely used in recent years. These efficient enzyme technologies include zinc finger nuclease (ZFN) technology, transcriptional activation-like effector nuclease (TALENs) technology, and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) 9 system (CRISPR/Cas9) technology. The CRISPR/Cas (Cas) system is a gene editing tool for DNA modification regulated by a short RNA and is a new type of genome editing tool that is faster, more efficient, and more accurate than the zinc finger nuclease and transcription activator-like effector nuclease. This article reviews the structure and function of the CRISPR/Cas system, and is aimed to outline the Cas9 design strategy, factors that affect the Cas9 gene editing efficiency, off-target detection and analysis methods, and especially the application in animal gene editing studies. Based on CRISPR/Cas9 gene editing has been successfully implemented in a variety of animals, and it is expected to become a new feasible way to establish animal models and study disease prevention in veterinary science and research.
Introduction
Gene knockout technology refers to a technique that inactivates or deletes a specific gene in a body through a certain pathway. In the early 1980s, the success of embryonic stem cell isolation and in vitro culture laid the technical foundation for gene knockout. Then in 1985, the existence of homologous recombination (HR) was first confirmed, which laid the theoretical foundation for gene knockout in mammalian cells [1,2]. In order to edit genes, traditional homologous recombination techniques that target specific alleles are used. However, this method generally has the disadvantages of low efficiency and high labor cost, which seriously restricts basic research and clinical application [3]. This requires scientists to explore more concise and efficient gene editing techniques.
RNA interference (RNA interference, RNAi) is a widely used genetic method for study the gene function of mammalian cells, and can also be used for RNA silencing of fungi [4]. It has the advantages of simple operation and obvious effects. However, RNAi could not act on all genes and certain cell types (such as neurons) [5,6], but also has the disadvantages of position effect, temporaryity and incomplete knockout. Therefore, gene editing techniques such as zinc finger nuclease technology [7], transcriptional activation-like effector nuclease technology [8], and clusters of regularly spaced short palindromic repeat sequences systems have been widely developed and used in recent years [9], which have revolutionized the contribution of gene editing. Initially, the CRISPR/Cas system cannot be applied to humans and animals. After its transformation, it has been widely used as a nuclease-based gene editing technology in animals [10], and this study mainly reviews the research progress of CRISPR in veterinary science and research.
Gene structure and action mechanism of CRISPR/Cas9 system
Gene Structure of CRISPR/Cas9 System: CRISPR/Cas9 is a new gene editing technology discovered in recent years. Currently there are three CRISPR/Cas systems (I, II, III), and the Cas9 nuclear protein enzymes mainly constitute and express Type II CRISPR/Cas systems. Each system contains a cluster of CRISPR-related genes, non-coding RNA, and a unique array of positive repetitive elements, in which the most common Cas9 is S. pneumoniae (sp-Cas9) system, the Type II CRISPR/Cas9 system. The CRISPR/Cas9 system requires at least 3 components: a CRISPR-related nuclease, a specific CRISPR RNA (CRISPRRNAs, crRNA) and a trans-activated CRISPR RNA (TransactivatingcrRNA, tracrRNA) [11]. In order to streamline this technique, the researchers designed a single guide RNA (gRNA) that replaces the crRNA-tracrRNA complex, which could direct the Cas9 nuclease to the targeted target site to cleave the double-stranded DNA [12]. In addition, there is a structure called tracrRNA tail on tracrRNA, which is beneficial to enhance the expression of Cas9 nuclease [13]. CRISPR is a series of short palindrome repeats that are separated by one another. The sequence is 21-48 bp in length. These repeats often produce hairpin structures, and the number of repeats of hairpin structures can reach 200 times or more. Each repetitive sequence is separated by a short repetitive sequence that is similar in structure to an exogenous DNA target, called the protospacer, which determines the type of CRISPR system and the recognition site of the target gene [14]. Within the DNA target, each typical spacer region is always adjacent to the protospace radjacent motif (PAM), and PAM can be varied according to the specific CRISPR system. There are three types of commonly used PAM, which is NGG, NAG and NNGG, respectively [15]. The Cas9 protein contains two nuclease domains, one is the HNH domain and the other is the RuvC-like domain. The HNH nuclease domain and the RuvC-like nuclease domain cut a single strand of the target DNA strand, respectively. And such single-stranded cleavage is prone to mutations, possibly due to the presence of HNH and RuvC [16].
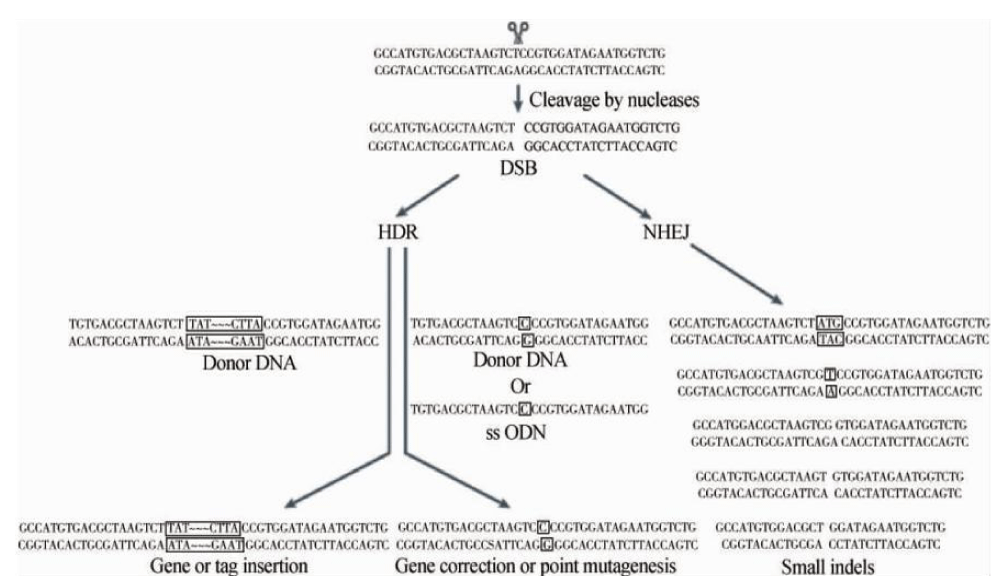
The action mechanism of CRISPR/Cas9 System: As shown in figure 1, the PAM sequence causes recognition of the Cas9 protein, allowing the single-stranded guide RNA linked to the tracrRNA to recognize the target sequence site, ensuring that the Cas9 protein and the genome stably binding, causing cleavage of the target site (usually the -3 and -4 upstream of the PAM), thereby producing the DNA double-strand breaks (DSB) [17]. DNA double-strand breaks cause non-homologous end joining or homologous orientation repair, and each single strand is repaired by a highly accurate base excision repair mechanism. The CRISPR/Cas9 system can perform efficient knockout and knock-in editing of the target gene simultaneously [18]. As shown in figure 2, after the target fragment cutted, the effect of knockout was achieved by the insertion or the missing mutation through the NHEJ repair mechanism. Knock-in transforms exogenous and functional genes into homologous sequences of genes and repairs them by homology-directed repair (HDR) to perform knock-in or point mutations for expressing the inserted gene in cells. Performing Knockout and Knock-in efficiently at specific sites simultaneously is the feature of Cas9 system [19], and it is only necessary to change the exogenous gene on the donor vector.
The design strategy of Cas9 system
Sequences design of target DNA: At present, it is still not possible to determine the exact criteria for designing target sequences [20], but according to some conventional criteria, select better target sites among many target sites is suitable. Currently designing targeting sequences of on-line software are https://chopchop.rc.fas.harvard.edu/, https://rgenome.net/, https://zifit.partners.org/ZiFiT/Choice-Menu.aspx, https://www.e-crisp.org/E-CRISP/de sign crispr.html, https://crispr.mit.edu/ [21]. In these online softwares, the main function of the first two is to find the target sequence. Afterwards, manual screening is required and the result is more reliable. This software, https://rgenome.net/, can be used to find target sequences and design guide RNAs and it can also set the required number of base mismatches.
Single-chain guide RNA design: The sgRNAs from 5’ to 3’ are DNA complementary regions, crRNA and tracrRNA, respectively, in which the design of the DNA complementary region has a crucial influence on the target efficiency [22]. The sgRNA has a 12-base unique sequence adjacent to the upstream of the PAM and is called the “seed region”, which has a greater effect on the recognition of mismatched target sites [23]. A certain number of mismatches between the sgRNA and the target fragment can be tolerated, especially if these mismatches are far away from PAM, so it is important to note the position of the mismatched sequence when designing sgRNA. The general criterion for designing sgRNAs for genomes is that a maximum of 1 mismatch is allowed at the 3’ end of the 12 base seed region immediately adjacent to the PAM sequence (5’-NGG-3’) [24]. In general, the 12-base sequence close to PAM should be designed strictly following the principle of base complementation, and ensured that there were no mutations or mismatches in the sequence [25].
Factors affecting the efficiency of Cas9 targeting: When designing the RNA, the GC content is generally 45% to 60%. Exceeding this range will affect the targeting efficiency of Cas9 and C base enrichment. The enrichment of C bases makes it easy to target DNA methylation sites, and trying to avoid methylation sites could help reduce epigenetic mutations [26]. However, it is contradictory that some studies have pointed out that methylation only affects ZFNs and TLAENs, and does not affect the cutting efficiency of Cas9. The DNA polymerase I hypersensitive site (DHS) belongs to a state of chromatin structure, which can significantly improve the cell-specific prediction of the transcription factor binding site. Designing the targeting sequence in DHS can improve the efficiency of gene editing [27]. In addition, when selecting the target site, try to select the exon region instead of the intron region because the intron has little meaning for the translation product and is more susceptible to mutation than the exon.
Application of CRISPR/Cas9 system in animal genetic editing research
Gene knockout: As early as in 2013, the CRISPR/Cas9 technology was performed for gene knockout on cell lines by scientists. Using target site-specific RNA, Cas9 nuclease was introduced to the target site of the genome to cleave and cause mutations. Then Cas9 gene targeting technique using mouse as an animal model has been reformed as a “one-step method” that enables multiple genome editing [28]. Niu Y et al., selected two target genes, Ppar-γ and Rag1, and injected Cas9 mRNA and single-stranded guide RNA into the fertilized eggs of cynomolgus monkeys. At the same time, these two genes were targeted in one step, and no off-target phenomenon was detected in the whole gene analysis and detection [29]. Finally, the genetically modified transgenic cynomolgus monkey was successfully obtained. Wang H et al., reported mutations caused by CRISPR/Cas9 technology in the Tet (Ten-eleven translocation family members) gene could cause multiple tumors, especially hematopoietic system tumors. The researchers co-injected Cas9 mRNA and sgRNA into mouse fertilized eggs for knocking out Tet1 and Tet2 by Microinjection method, and the knockout efficiency was verified to be about 80%. The experiment successfully produced a small biallelic mutation rats, and for the first time, knock out two endogenous genes simultaneously in animals [30].
In order to promote the veterinary science and research, CRISPR/Cas9 technology was employed to improve animal strains. Zhou X et al used Cas9/sgRNAs to knock out Parkin2 and PINK1 genes (PTEN-induced putative kinase 1) on porcine fetal fibroblasts and cloned mutant cells into donors for somatic cell transplantation to produce homozygous transgenes pig. The myostatin encoded by Myostatin (myostatin) inhibits muscle differentiation and growth [31]. Chen F et al used the CRISPR/Cas9 system to knock out the JH (Joining chain) region of the porcine immunoglobulin M (IgM) heavy chain gene, which plays a crucial role in the development and differentiation of the immune system. With the support of somatic cell nuclear transfer technology, the efficiency of gene knockout was 53.3% after transfection of pig embryo fetal fibroblasts with IgM antibody Cas9 plasmid, of which 25% of positive clones had biallelic modification, which is more efficient than the traditional homologous recombination [32]. The researchers used CRISPR/Cas9 technology to microinject the CRISPR/Cas9 mRNA of the specific MSTN gene into the cytoplasm of ovine fertilized eggs, and the results showed that the embryo development mutation rate reached 50% [33]. In goats, the efficiency of knocking out MSTN and FGF5 genes in fibroblasts was close to 60%, and only 15% and 21% of animals survived after knocking out MSTN and FGF5 genes in 98 experimental animals, respectively. After both double gene modification, and the result showed 10% of animals survived. These studies suggest that the CRISPR/Cas9 system can be used as an effective gene editing tool for breeding new varieties of animal traits and breeding for disease resistance [34]. By injecting the Cas9 mRNA and sgRNA of the IL2RG and Rag1 genes into the cytoplasm of prokaryotic embryos, bi-allelic knockout rabbits can be obtained and the efficiency up to 100%, while knocking out 3 genes (IL2RG, RAG1, and RAG2) and 5 genes (IL2RG, RAG1, RAG2, TIKI1 and ALB), the efficiency could reach 33.3% [35]. N. Véron et al used live point punching technology to effectively knock out the transcription factor PAX7 (Paired box7) in chicken embryonic stem cells, resulting in mosaic gene mutations in wild-type multicellular animals, and loss of related functions of chicken embryonic stem genes [36]. Moreover, the Cas9 technology also enables efficient multigene modification on zebrafish, and the crRNA-tracrRNA-Cas9 protein complex visualizes endogenous gene expression, this is the first breakthrough in the cold water animal model [37].
Gene knock-in: Studies have found that if homologous DNA is provided, exogenous sequences could be knocked into specific target sequence of zebrafish embryos with an insertion efficiency of 3.5% to 15.6%. The potential off-target sites is only 1.1% to 2.5%, reflecting the specificity of the Cas9 system [38]. To date, few articles have reported about the use of the Cas9 system to edit bovine genomes, probably because of the longer gestation cycle and the off-target nature of Cas9 [39]. In 2016, the researchers used the NHEJ pathway to efficiently integrate a 4.6 kb promoter-less vector into the GADPH gene locus, with knock-in rates of 20% and 1.7% on human cells and embryonic stem cells, respectively, and the NHEJ approach is demonstrated more efficient than the HDR approach [40], however, the cytotoxicity problems that may be caused by the NHEJ pathway have not been evaluated.
Conclusion
The Cas9 system is a unique mechanism for microbial self-protection, which is designed to prevent the invasion of foreign microorganisms. It is a faster and more efficient gene editing tool after ZFN and TALEN. This technology aims to improve the recognition and binding ability of specific sequences and the efficiency of enzyme digestion. More and more studies have improved the gene edit possibility in different species by using Cas9 system. Since the PAM (5’-NGG-3’) of Cas9 nuclease is very short and can be found in almost any species, this solves the problem of cross-species in gene editing tools. Efficient, low time cost, multi-site gene editing on a genome, and basically no species restrictions are the advantages of Cas9 gene editing technology. Cas9 gene editing technology also has some shortcomings, such as easy off-target and potential target sites, gene mutations and too many factors affecting target efficiency. Gene mutations generated during gene editing are side effects of the CRISPR/Cas9 system targeting system, and CRISPR/Cas9 nucleases may bind to unexpected sites, leading to genetic mutations at certain sites, which are aspects of the Cas9 system that need improvement.
Researchers are also working to continuously explore and optimize the research of the CRISPR system to make it faster, easier, and better. Slaymaker IM et al found a protein smaller than Cas9 nuclease in S. aureus, which is more powerful in easier access to mature cells and easier to binding ability to the vector, and then the new eSpCas9 gene editing system was transformed, which not only reduced the off-target efficiency, but also continued efficient and high-accuracy target efficiency [41]. It is believed that with the continuous optimization and improvement of CRISPR/Cas9 gene editing technology, it plays an important role in animal new breed cultivation, disease resistance breeding and disease research and development in biomedical fields.
This study was supported by the National Natural Science Foundation of China (81672913), and the Maternal and Child Health Research Project of Jiangsu Province (F201604). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
- Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS (1985) Insertion of DNA sequence into human chromosomal. β-globin locus by homologous recombinantion. Nature 317: 230-234. Link: https://tinyurl.com/yae62p8z
- Capecchi MR (1989) Altering the genome by homologous recombination. Science 244: 1288-1292. Link: https://tinyurl.com/ydhu7bjz
- Bartosova Z, Krejci L (2016) Nucleases in homologous recombination as targets for cancer therapy - FEBS Letters. Febs Letters 588: 2446-2456. Link: https://tinyurl.com/y84ru725
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, et al. (2003) Expression profiling reveals off-target gene regulation by RNAi. Nature Biotechnology 21: 635-637. Link: https://tinyurl.com/y9yrajf2
- Asikainen S, Vartiainen S, Lakso M, Nass R, Wong G (2005) Selective sensitivity of Caenorhabditis elegans neurons to RNA interference. Neuroreport 16: 1995–1999. Link: https://tinyurl.com/ydem7q4t
- Qadota H, Inoue M, Hikita T, Köppen M, Hardin JD, et al. (2007) Establishment of a tissue-specific RNAi system in C. elegans. Gene 400: 166–173. Link: https://tinyurl.com/yb5j9vpf
- Jo YI, Kim H, Ramakrishna S (2015) Recent developments and clinical studies utilizing engineered zinc finger nuclease technology. Cellular & Molecular Life Sciences Cmls 72: 3819-3380. Link: https://tinyurl.com/ydyex64d
- Moore FE, Reyon D, Sander JD, Martinez SA, Blackburn JS, et al. (2012) Improved somatic mutagenesis in zebrafish using transcription activator-like effector nucleases (TALENs). Plos One 7: e37877. Link: https://tinyurl.com/y8enoc7r
- Rezzonico F, Smits THM, Duffy B (2011) Diversity, evolution, and functionality of clustered regularly interspaced short palindromic repeat (CRISPR) regions in the fire blight pathogen Erwinia amylovora. Appl Environ Microbiol. 77: 3819. Link: https://tinyurl.com/y9cwj9s6
- Shrock E, Güell M (2017) Chapter Six - CRISPR in Animals and Animal Models. Prog Mol Biol Transl Sci 152: 95-114. Link: https://tinyurl.com/ycga2eo5
- Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR-Cas9 for Genome engineering. Cell 157: 1262-1278. Link: https://tinyurl.com/ycfnkhcg
- Pelletier S, Gingras S, Green DR, et al. (2015) Mouse genome engineering via CRISPR-Cas9 for study of immune function. Immunity 42: 18-27. Link: https://tinyurl.com/yb6tdd56
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, et al. (2013) DNA targeting specificity of RNA-guided Cas9 nucleases. Nature Biotechnology 31: 827. Link: https://tinyurl.com/ybyylzcj
- Doudna JA, Charpentier E (2014) The new frontier of genome engineering with CRISPR-Cas9. Science 346: 1258096. Link: https://tinyurl.com/ybtn9k7g
- Xiao A, Cheng Z, Kong L, Zhu Z, Lin S, et al. (2014) CasOT: a genome-wide Cas9/gRNA off-target searching tool. Bioinformatics 30: 1180-1182. Link: https://tinyurl.com/ya9qxnql
- Cong L, Ran FA, Cox D, Lin S, Barretto R, et al. (2013) Multiplex genome engineering using CRISPR/Cas system. Science 339: 819-823. Link: https://tinyurl.com/y85oow8e
- Karvelis T, Gasiunas G, Siksnys V (2017) Methods for decoding Cas9 protospacer adjacent motif (PAM) sequences: A brief overview. Methods 121-122: 3-8. Link: https://tinyurl.com/yd8aq7xj
- Dianov GL, Hubscher U (2013) Mammalian base excision repair: the forgetten archangel. Nucleic Acids Res 41: 3483-3490. Link: https://tinyurl.com/y88mfb7c
- Kim H, Kim JS (2014) A guide to genome engineering with programmable nucleases. Nat Rev Genet 15: 321-334. Link: https://tinyurl.com/ydef9xfh
- Carroll D (2013) Staying on target with CRISPR-Cas. Nat Biotechnol 31: 807-809. Link: https://tinyurl.com/y85j3vtk
- Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E (2014) CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res 25: 1-7. Link: https://tinyurl.com/yahbyxqz
- Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P,et al. (2011) The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res 39: 9275-9282. Link: https://tinyurl.com/yaem73on
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA (2013) RNA-guided editing of bacterial genomes using CRISPR-Cas system. Nat Biotechnol 31: 233-239. Link: https://tinyurl.com/yb48dwsj
- Jinek M, East A, Cheng A, Lin S, Ma E, et al. (2013) RNA-programmed genome editing in human cells. Elife 2: e00471. Link: https://tinyurl.com/y8q22me9
- Cho SW, Kim S, Kim JM, et al. (2013) Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31: 230-232. Link: https://tinyurl.com/yb97yaol
- Wiles MV, Qin W, Cheng AW, Wang H (2015) CRISPR-Cas9-mediated genome editing and guide RNA design. Mamm Genome 26: 501-510. Link: https://tinyurl.com/y9684ncl
- Gusmao EG, Dieterich C, Zenke M, Costa IG (2014) Detection of active transcription factor binding sites with the combination of DNase hypersensitivity and histone modification. Bioinformatics 30: 3143-3151. Link: https://tinyurl.com/y8rvtnxt
- Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, et al. (2013) One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 153: 1370-1379. Link: https://tinyurl.com/ydbjuk6g
- Niu Y, Shen B, Cui Y, Chen Y, Wang J, et al. (2014) Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting on one-cell embryos. Cell 156: 836-843. Link: https://tinyurl.com/ybdwskpz
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, et al. (2013) One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153: 910-918. Link: https://tinyurl.com/y7rhsujj
- Zhou X, Xin J, Fan N, Zou Q, Huang J, et al. (2015) Generation of CRIPR/Cas9-mediated gene-targeted pigs via somatic cell nuclear transfer. Cell Mol Life Sci 72: 1175-1184. Link: https://tinyurl.com/ybk5xacp
- Chen F, Wang Y, Yuan Y, Zhang W, Ren Z, et al. (2015) Generation of B cell-dificient pigs by highly efficient CRISPR/Cas9-mediated gene targeting. J Genet Genomics 42: 437-444. Link: https://tinyurl.com/y7qjnd2p
- Crispo M, Mulet AP, Tesson L, Barrera N, Cuadro F, et al. (2015) Efficient generation of myostatin knock-out sheep using CRISPR/Cas9 technology and microinjection into zygotes. PLos One 10: 366-375. Link: https://tinyurl.com/y76kwln6
- Wang X, Yu H, Lei A, Zhou J, Zeng W, et al. (2015) Generation of gene-modified goats targeting MSTN and FGF5 via zygote injection of CRISPR/Cas9 system. Sci Rep 5: 13878. Link: https://tinyurl.com/yabldanr
- Yan Q, Zhang Q, Yang H, Zou Q, Tang C et al. (2014) Generation of multi-gene knockout rabbits using the Cas9/gRNA system. Cell Regen 3: 12. Link: https://tinyurl.com/yatom5vq
- Veron N, Qu Z, Kipen PA, Hirst CE, Marcelle C (2015) CRISPR mediated somatic cell genome engineering in the chicken. Dev Biol 407: 68-74. Link: https://tinyurl.com/y7jy8glb
- Hrucscha A, Krawitz P, Rechenberg A, Heinrich V, Hecht J, et al. (2013) Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development 140: 4982-4987. Link: https://tinyurl.com/yagdmcvb
- Kotani H, Taimantsu K, Ohga R, Ota S, Kawahara A (2015) Efficient multiple genome modification induced by the crRNA, tracrRNA and Cas9 protein complex in zebrafish. PLos One 10: 128-134. Link: https://tinyurl.com/y7fj6h5e
- Wang Z (2015) Genome engineering in cattle: recent technological advancements. Chromosome Res 23: 17-29. Link: https://tinyurl.com/y7fv4eyb
- He X, Tan C, Wang F, Wang Y, Zhou R, et al. (2016) Knock-in of large reporter genes in human cells via CRISPR/Cas9-induced homology-dependent and independent DNA repair. Nucleic Acids Research 44: e85-e85. Link: https://tinyurl.com/ya6otxr7Slaymaker IM, Gao I, Zetsche B, Scott DA, Yan WX, et al. (2016) Rationally engineered Cas9 nucleases with improved specificity. Science 351: 84-88. Link: https://tinyurl.com/y83qu7av
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
