International Journal of Sexual and Reproductive Health Care
Strategies to fight COVID-19: Beyond the difference between SARS-CoV-2 and Influenza virus
Huihui Lin and Hequan Li*
Cite this as
Lin H, Li H (2022) Strategies to fight COVID-19: Beyond the difference between SARS-CoV-2 and Influenza virus. Int J Sex Reprod Health Care 5(1): 016-029. DOI: 10.17352/ijsrhc.000034Copyright Licence
© 2022 Hequan Li, MD, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Background: Since the outbreak of COVID-19 emerged in Wuhan, China, in December 2019, the epidemic has spread worldwide and posed a great threat to society. Despite great achievements in COVID-19 research, few studies have focused on the similarities and differences between SARS-CoV-2 and influenza viruses.
Results: Through a review of the literature on SARS-CoV-2 and influenza viruses, we found that influenza occurs every year, and influenza pandemics occur irregularly. The uncomplicated human influenza viruses primarily affect the larger airways and rarely the alveoli. However, SARS-CoV-2 mainly involves the deep airways and lungs and can cause DAD, leading to severe hypoxemia. In general, SARS-CoV-2 is no less infectious than the influenza virus. However, its destructive power to the lungs is no less than the avian influenza virus. There is currently no clinical vaccine and specific inhibitor against SARS-CoV-2.
Conclusions: SASR-CoV-2 damages lung function more severely than the influenza virus, with higher morbidity, mortality, and severe disease rates. Controlling the source of infection, cutting off the route of transmission, and protecting susceptible populations are critical to the fight against SARS-CoV-2.
Introduction
As of May 27, 2022, data from the World Health Organization (WHO) showed 525,467,084 confirmed cases of COVID-19 worldwide [1]. Furthermore, as of May 22, 2022, almost one billion people in lower-income countries remain unvaccinated. Only 57 countries have vaccinated 70% of their population-almost, all high-income countries [2]. That means that the whole world is in battle with SARS-CoV-2. Both COVID-19 and influenza are characterized by fever and respiratory symptoms [3,4]. According to the Law of the people’s Republic of China on the Prevention and treatment of Infectious Diseases, COVID-19 belongs to Class B infectious diseases and is managed in accordance with Class A. Although human infection with highly pathogenic avian influenza also belongs to class B infectious diseases, influenza belongs to class C. Hence, the prevention and control measures and management systems are different. Presently, there have been many studies comparing COVID-19 to previous influenza pandemics. To understand how to contain the spread of SARS-CoV-2 more effectively, we examined the similarities and differences between SARS-CoV-2 and influenza viruses and discussed six aspects of outcomes, clinical features and treatment.
Transmission
Influenza virus: At present, the available evidence supports the important role of all routes of transmission (droplet, aerosol, and contact) in the spread of influenza. The problem is their relative importance, which will depend on the set of circumstances acting at a given time [5], while the avian influenza virus spreads inefficiently from person to person [6].In the United States, about 5% to 20% of the population gets the flu [7,8] and an estimated 3000 to 49,000 people die of influenza each year [9], then the case fatality rate (CFR) is much less than 1%, and the mortality rate is even lower. Between 2010 and 2016, an average of about 8% of Americans contracted the flu each season. And the median value of children 0-17 is more than twice that of adults over 65. [8]. And the Centers for Disease Control and Prevention (CDC) points out that anyone can be infected with influenza. People over the age of 65, people of any age with certain chronic diseases, pregnant women, and children under the age of 5 are at high risk of severe flu-related complications [10]. Actually, the infectivity and mortality of influenza are closely related to specific virus types and epidemic subtypes. For example, the course of H1N1 subtype is worse than that of H3N2 subtype, and most influenza cases need hospitalization with high mortality [11].
SARS-CoV-2
Nowadays, SARS-CoV-2 has spread worldwide as an Omicron variant [12]. It is certain that SARS-CoV-2 can be transmitted from person-to-person through droplets produced by coughing or sneezing or through close contact [13]. SARS-CoV-2 can also be transmitted in the air through aerosols formed during medical treatment [14]. And viral nucleic acid can be found in patients’ feces and rectal swabs, making the argument that SARS-CoV-2 spread to the mouth through feces is convincing [15]. In addition to this, the deposition of virus-laden aerosols may contaminate objects (eg, fomites) and lead to human transmission events [16]. SARS-CoV-2 can directly infect human renal tubules and may cause urine transmission [17]. Therefore, respiratory droplets and close contact are the main ways, and SARS-CoV-2 may be transmitted by aerosols if they are exposed to high concentrations of aerosols for a long time in a closed environment. Because SARS-CoV-2 can be isolated from feces and urine, attention should be paid to the aerosol or contact transmission of environmental pollution caused by feces and urine [18]. According to the Report of the WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19), most laboratory-confirmed cases (77.8%) are between 30 and 69 years old. About 80% of cases are mild and common, 13.8% are severe, and 6.1% are critical. People at high risk of severe illness and death are those over the age of 60 and those with basic diseases [19].And as of May 27, the crude CFR of COVID-19 globally is about 1.20% [1]. In short, current COVID-19 epidemic is serious, the proportion of COVID-19 severe/critical patients is not low, and the dead are mostly accompanied by underlying diseases.
In terms of routes of transmission, both influenza virus and SARS-CoV-2 can be transmitted through droplets, contact, and aerosols, but the fecal and urethral transmission of SARS-CoV-2 cannot be ruled out. Children are the most susceptible population to catching influenza, while child cases of COVID-19 seem less [19]. In order to compare infectivity, we use a parameter, the basic reproduction number (R0). R0 refers to the average expected number of new cases that a case will produce in other uninfected people during its infectious period [20]. The R0 of SARS-CoV-2 is estimated to be 2.2-2.6 [21]. Noteworthy, from the method of calculating R0 proposed by Ndaïrou et al, the transmission coefficient due to super-spreaders is positively correlated with R0 [22]. Li, et al elaborated on a COVID-19 super-spreader who was misdiagnosed before the operation to show that the existence of super-spreaders greatly contributes to the spread of SARS-CoV-2 [23]. And the occurrence of super communication events promotes spread [24]. While the median R0 value of seasonal influenza is 1.28, and that of several influenza pandemics over the past 100 years is 1.46 [25]. Furthermore, SARS-CoV-2 can display transmissibility and lethality of a particularly deadly pandemic virus, second only to the pandemic influenza virus of 1918 [26,27]. Overall, except for the influenza pandemic, SARS-CoV-2 is more contagious, with higher CFR (Table 1).
Pathogenesis
Influenza virus: Influenza virus (IV) is encapsulated by two kinds of surface glycoprotein hemagglutinin (HA) and neuraminidase (NA). HA is the main antigen of neutralizing antibodies, and the infectivity of influenza depends on the cleavage of HA by specific host proteases [4,28].
The influenza virus invades the host by recognizing the sialic acid of surface molecules through HA proteins. The HAs of human influenza viruses preferentially bind to receptors molecule of α-2,6-linked sialic acid while avian and equine influenza viruses bind to α-2,3-linked sialic acid preferentially [29]. The distribution of these receptors in different tissues reflects the target cells and attack tendency of the virus [30]. The main receptors distributed on the epithelial cells of the nasal mucosa, paranasal sinuses, pharynx, trachea, and bronchi are α-2, 6-linked sialic acid, which is also expressed on epithelial cells of the terminal and respiratory bronchioles. And α-2,3-linked sialic acid is distributed on the surface of non-ciliated cuboidal bronchiolar cells at the junction between the respiratory bronchiole and alveolus and type II alveolar epithelial cells(AT2s) [6]. Therefore, human-derived viruses bound extensively to epithelial cells in the bronchi and, but less widely to alveolar cells, while avian viruses do the opposite.
Influenza virus invades airway epithelial cells and replicates, triggering the release of cytokines and chemokines, which can promote the recruitment of blood-derived inflammatory cells. That is, influenza can change the composition of the monocyte-derived alveolar macrophages (AMs) population and recruit more AM populations, in which BM-derived cells can also gather in the lungs. This kind of cell population is likely to be Ly6Chi monocytes. The migration of this kind of cells from bone marrow is mediated by CC-chemokine receptor 2(CCR2) and regulated by bone marrow stromal cells, which perceive circulating microbial molecules and produce CC‑chemokine ligand 2(CCL2). In addition, influenza-experienced alveolar epithelial cells can also produce CCL2, to facilitate the trafficking of monocytes. This indicates that in addition to mediating the recruitment of monocytes from bone marrow to the bloodstream, CCR2 help recruits monocytes to the infected lungs or lymph nodes that drain infected sites, trigger virus-specific CD8+T cells, thus limiting further growth and invasion of the virus, and starting viral clearance [31,32]. The above contents are consistent with Lin’s claim that most inflammatory cells in influenza-infected lungs come from CCR2+ monocytes, which are the predominant cause of lung immunopathology and mortality [33]. Therefore, the activation of AMs and the initiation of inflammatory cell recruitment affect the regression of inflammation and virus clearance, causing virus-induced pathology and mortality [33,34]. Meanwhile, the recruitment of CCR2-dependent monocytes is at the core of prolonged antibacterial protection after influenza virus clearance and clinical rehabilitation. At a month post-influenza, recruited macrophages transcriptionally resemble monocytes and provide prolonged antibacterial protection through increased production of IL-6 and other mechanisms. However, only monocytes recruited during the period of acute influenza sustained a phenotype of increased IL-6 production. After 2 months of influenza, the recruited AM population of the lungs is abundant, but transcriptionally and functionally similar to resident AMs and no longer provides protection. Thus, it can be seen that respiratory infection can change the AM population through monocyte-derived recruited cells and thus constantly alter pulmonary immunity [31].
As for influenza virus-specific CD4+ and CD8+T cells, they can recognize a variety of viral antigens, including HA and internal proteins, in which the surface hemagglutinin (HA) expressed on the surface of influenza virus mainly induces CD4+response, while CD8+ response is mainly targeted at virus internal proteins, resulting in protective immunity and cross-reaction with a variety of influenza viruses, which are important in resisting IV infection [35]. The pre-existence of CD4+T cells is associated with lower virus shedding and less severe disease outcomes. Moreover, CD4+T cells can secrete IFN-γto promote antiviral immune response, obtain cytotoxic functions, and clear infected cells through cytokine-mediated and perforin/granzyme-dependent mechanisms [36-41]. CD4+T cells are also important to induce and maintain virus-specific CD8+T cell-mediated immunity [42-45]. On the other hand, CD8+ cytotoxic T lymphocytes (CTL) induce apoptosis through the release of perforin and granzyme B and the interaction of Fas/Fas ligands, to identify and kill infected cells and thus preventing more progeny virus from being generated. Besides, activated CD8+T cells can also produce pro-inflammatory cytokines, such as IFN-γ, which inhibit viral replication[37]. It is worth mentioning that the production of virus-neutralizing antibodies by B cells is considered to be the best correlate of protection against IV infection [46]. Overall, infected cells can be killed through viral cytopathic effect and immune response to virus infection can also result in cell death and tissue damage [47].
SARS-CoV-2
SARS-CoV-2 has at least four classical structural proteins: Spike (S) protein, Envelope (E) protein, Membrane (M) protein, and Nucleocapsid (N) protein [48].
SARS-CoV-2 is structurally similar to SARS-CoV and has a similar receptor-binding domain structure (RBD), so they may share similar pathogenesis [49]. There is overwhelming evidence that the main mechanism of virus invasion into host cells is endocytosis, and clathrin-dependent endocytosis and cathepsin-mediated S protein cleavage are key steps for viral entry and infection [50]. Coronavirus mainly binds to specific cell surface receptors through the functional unit S1 of RBD, and S2 fuses the cell and virus membrane to make the virus genome enter the host cell. The RBD of SARS-CoV-2 has a higher affinity binding to its receptor than SARS-CoV [49]. Therefore, SARS-CoV-2 may endocytosis through binding to angiotensin-converting enzyme 2(ACE2) on the surface of host cells, then enter the target cells and eventually cause infection [50,51]. ACE2 was expressed in AT2s, cardiomyocytes, arteriovenous endothelial cells, arterial smooth muscle cells, renal tubular epithelial cells, bladder urothelial cells, esophageal epithelial cells, and stratified epithelial cells, bile duct cells, absorptive intestinal cells of ileum and colon [49,52-55]. Notably, key mutations in the RBD of SARS-CoV-2 Spike create additional close contacts with ACE2, which correlate with higher binding affinity and possibly increased infectivity [51]. After invading host cells, coronavirus replicates actively and triggers inflammatory responses through various signal pathways. Firstly, cytotoxic T lymphocytes and NK cells in COVID-19 patients are necessary to trigger appropriate anti-viral responses [56], in which CD8+T cell plays a crucial role in the pathogenesis [21]. Once into the tissue cells, viral peptides are delivered to CD8+ cytotoxic T cells through the class I major histocompatibility complex (MHC) proteins. After being activated, CD8+T cells develop virus-specific effector T cells and memory T cells, then CD8+ cytotoxic T cells can dissolve the tissue cells infected by the virus [36]. Moreover, type I IFN can enhance the cytotoxicity of CD8+T cells [57]. And the delayed kinetics of virus clearance is the trigger. The delayed-type I interferon response is important in the pathogenesis of SARS, which may result in early and rapid replication of the virus in the airway and alveolar epithelial cells, causing plenty of epithelial and endothelial cell apoptosis and vascular leakage. Besides, the activation of this signal pathway induces the widespread expression of the interferon-stimulating gene (ISG) and attracts inflammatory monocytes-macrophages (IMM), neutrophils, dendritic cells, and natural killer cells into the lungs [21,58,59]. Meanwhile, Th1 type response is the key to controlling SARS-CoV and may be true for SARS-CoV-2 [21]. In a short time, the virus and viral particles are recognized by antigen-presenting cells, and viral peptides are delivered to CD4+T cells through MHC-Class-II molecules [36]. Finally, proinflammatory cytokines and chemokines are greatly released [21]. The uncontrolled release of pro-inflammatory mediators suggests the existence of a “cytokine storm”, also known as cytokine release syndrome(CRS), a common immunopathological mechanism [58]. As mentioned before, SARS-CoV-2, like SARS-CoV, can increase the secretion of IL-1b, IFN- γ, IP-10, MCP-1, IL-4, and IL-10, etc [60], which may be related to a “cytokine storm”. The emergence of the “cytokine storm” causes the immune system of the body to respond quickly, killing the virus with suicidal attacks, while also causing damage to blood vessels and organs, tissues and cells [61-63]. In addition, many patients had lymphocytopenia [3], which may indicate the existence of functional exhaustion of cytotoxic lymphocytes, and the collapse of antiviral immune function contributes to the pathogenesis and severity of COVID-19.
In the early stage of SARS-CoV infection, active viral replication, viral-mediated down-regulation, and shedding of ACE2 and host antiviral response cause a primary inflammatory response. The occurrence of secondary inflammation begins with the production of adaptive immunity and the emergence of neutralizing antibodies (NAb). Although adaptive immunity and neutralizing antibodies can further reduce viral replication, they can trigger violent inflammation, resulting in serious tissue damage. When the neutralizing antibody activity produced by patients in the early stage of infection is lower than the optimal level, causing the virus not to be completely cleared, which may cause persistent viral replication and inflammation through antibody-dependent-enhancement(ADE) [64]. When the neutralizing antibody binds to S protein in vivo, it can bind to the FcR receptor expressed on monocytes/macrophages [65], causing secondary inflammatory reactions.
In a word, human influenza virus attachment in the trachea and bronchi was more abundant than in the bronchioles and alveoli. In alveoli, the virus attached more to type I pneumocytes than to AT2s and rarely to alveolar macrophages. However, the avian influenza virus is mainly attached to the alveoli (preferentially attached to AT2s) and bronchioles [66]. Excessive pulmonary inflammation and lung tissue injury during influenza virus infection are mainly due to the interactions of the virus with host cells, particularly those of the macrophage lineage. In the lungs, SARS-CoV-2 is mainly attached to the AT2s expressing ACE2 to trigger the cascaded amplified inflammation through various innate immune responses, T and B cell immunity, and antiviral neutralizing antibody responses (Table 2).
Laboratory examination
Influenza virus: Most patients with influenza/avian influenza have normal or slightly decreased leukocyte count and decreased lymphocyte count, which may be accompanied by changes in other non-specific indexes [67,68]. Of note, however, that lymphopenia is a clear risk factor for severe influenza and may contribute to the early differential diagnosis of influenza A/H1N1 pandemic [69,70]. And low levels of circulating CD8+T effector and central memory cells are related to the severity of influenza [71]. At present, the main methods to detect virus antigens are immunochromatography (IC) and direct/indirect immunofluorescence (DFA/IFA). The more sensitive etiological examination is nucleic acid amplification technology (NAAT), which is mainly used for nucleic acid detection, and includes end point PCR and reverse transcription-polymerase chain reaction (RT-PCR). However, the detection of influenza virus RNA or nucleic acid through these tests does not necessarily mean the detection of live viruses or ongoing viral replication [10,72]. Serological diagnosis of influenza is based on the detection of a four-fold or greater rise in specific antibody titers in paired serum samples, measured by hemagglutination inhibition test(HIA), enzyme immunoassay(EIA), complement fixation, and neutralization tests [4]. And serological tests of influenza infection require paired acute and convalescent sera. Since influenza infection generally represents reinfection, the detection of influenza-specific antibodies on a single serum sample cannot diagnose recent infection [73].
SARS-CoV-2
Studies demonstrate that the absolute value of lymphocytes in patients with COVID-19 is significantly decreased, especially in severe patients [3,74-77]. And the level of cytokines in ICU patients is higher than that in non-ICU patients [60]. Therefore, the decrease in lymphocyte counts and the increase in CRP can be used as reference indexes to diagnose COVID-19, while CD3+T, CD4+T, and CD8+T lymphocytes count and the level of cytokines are related to the severity of the disease and can be used to predict the severity [76,77]. Studies indicate that CD4+ and CD8+ T cells in acute COVID-19 patients(≥55 years old) have a lower capacity to produce IFN-γ and IL-2, and impaired T cell activation of dendritic cells (DCs) may compromise optimal adaptive immune responses [78]. Notably, CD8+T cells tended to be an independent predictor for COVID-19 severity and treatment efficacy [79]. The serological diagnosis of COVID-19 requires that the serum SARS-CoV-2 specific IgM antibody and IgG antibody are positive, or specific IgG antibody changes from negative to positive, or the level in the convalescent stage is four times higher than that in the acute stage. The detection of serological antibodies can be one of the diagnostic bases for suspected cases [18]. Molecular diagnosis of SARS-CoV-2 by RT-PCR in respiratory specimens is considered the gold standard method. There are also some rapid antigen test kits available for screening symptomatic and asymptomatic patients and their contacts [80].
Imaging examination
Influenza virus: The imaging findings of mild influenza may have no obvious abnormality, or only pulmonary texture thickening and disorder (Figure 1a) [81]. The most common imaging manifestation of influenza complicated with pneumonia is interstitial changes (Figure 1b) [11,82], mostly bilateral reticulonodular opacity with or without focal consolidation, usually in the lower lobes and peribronchovascular zone of bilateral lungs [83,84]. Oliveira et al found that the most common radiological abnormalities of seasonal influenza were bilateral diffuse interstitial/alveolar infiltration [85]. And the most common pulmonary X-ray findings of severe H1N1 patients in ICU are ground-glass opacities(GGO) and consolidations, followed by reticular structures, which are mainly seen in bilateral lungs, lower and middle lung fields, and often involve more than 3 lung regions. The more severe patients are, the higher the incidence of GGO and consolidation, and the more lung areas involved [86,87].In addition, the main imaging manifestations of avian influenza are GGO and consolidations (Figure 1c), which can be focal, multifocal, or diffuse, and are changing fast, spreading rapidly, and absorbing slowly, with or without centrilobular nodules, pseudocavitation, emphysema, and lymphadenopathy. Of note, compared with severe influenza pneumonia, the extent and degree of lung imaging of H7N9 avian influenza are more severe [68,88-90]. Briefly, the main imaging manifestation of influenza pneumonia is bilateral interstitial/alveolar infiltration, with or without consolidation, which can display GGO, and is related to the severity of the disease and can be used to predict the prognosis (Figure 1), but the normal imaging findings in the early stage cannot rule out the possibility of influenza or poor prognosis [87,91,92].
SARS-CoV-2
The typical lung images of COVID-19 showed bilateral patchy shadows and GGO [3,75-77]. The most common manifestations in the initial stage of COVID-19 were simple GGO, GGO with reticular and/or interlobular septal thickening, and GGO with consolidation. Most of these manifestations are patchy or nodular lesions, mainly involving interstitial lobules. The lesions were characterized by multifocal distribution in the middle and lower part of the lung and in the posterior part of the lung. Complete consolidation is relatively rare [77,93].
Notably, the proportion of involvement of both lungs and the lower lobe in the right lung in severe patients was higher than that in non-severe patients[76]. Their imaging findings showed bilateral diffuse consolidation shadow, which could progress to “white lungs” [94]. This shows that with the progress of the disease, the lesions can be fused into tablets, and pulmonary aggravation and repair can coexist [77]. Therefore, consolidation lesions can serve as a sign of disease progression or more serious diseases [93]. And there are some typical signs of COVID-19, such as the “pleural parallel sign”, “halo sign”, “reverse-halo sign” and vascular sign which are helpful for imaging diagnosis [94]. Besides, studies suggest that the prevalence of acute pulmonary embolism in patients with COVID-19 is high [95], so angiography can show bilateral main pulmonary arteries filling defects and peripheral GGO, and so on [96, 97].
In conclusion, CT findings of COVID-19 showed mixed and multiple types of pulmonary parenchyma and interstitial involvement, and typical images showed bilateral multiple patchy GGO and/or consolidation shadows (Figure 2). Most of the lesions were distributed along with the bronchovascular bundle or dorsolateral or subpleural [77,93,98,99]. Moreover, the imaging findings are related to the severity of the disease, which is helpful to judge the severity of patients and evaluate the prognosis [100].
Thus, in terms of imaging findings, COVID-19 is similar to those of avian influenza and more severe than influenza (Table 3).
Pathological findings
Influenza virus: Respiratory tract and lungs: The pathology caused by influenza viruses in humans depends on the virulence of the source of infection and host responses; Uncomplicated human influenza virus infection can cause transient tracheobronchitis, the early stages of the disease often demonstrate tracheobronchitis and bronchiolitis, airway walls congestion, monocytes infiltration, and epithelial cell degeneration. When the disease aggravates, parenchymal change shows typical diffuse alveolar damage (DAD) with hyaline membrane formation, accompanied by varying degrees of alveolar edema and/or hemorrhage, interstitial and lacunar infiltration, and vessel thrombosis. Repair-related changes, such as the proliferation of fibroblasts can be observed in the long course of the disease [83,104-106]. Furthermore, concrete pathological manifestations caused by influenza viruses also vary from type and subtype. Low-virulence viruses mainly cause inflammation, hyperemia, and necrosis of mucosal epithelial cells in larger airways (trachea, bronchus, and bronchioles), while high-virulence viruses(such as H5N1) tend to infect alveolar macrophages and pneumocytes, which can cause DAD [107].
Other organs
Studies show that there is viremia in human influenza virus infection, which may cause the brain, heart, and other organs to be affected, but this has yet to be studied[105, 108-110].On the other hand, when influenza virus infection causes acute respiratory distress syndrome(ARDS) to develop into multiple organ dysfunction syndromes (MODS), the liver, kidney, central nervous system, gastrointestinal, blood, and cardiac systems are most commonly affected. Therefore, when other organs/systems are involved, pathological changes can present encephalopathy, myocarditis, myositis or myopathy, and so on. Moreover, influenza virus RNA in inflamed myocardium can be identified in some cases [105,110-112].
All influenza viruses infect the respiratory epithelium from the nasal cavity to the bronchioles [107]. Non-fatal influenza virus infection mainly involves the upper respiratory tract and trachea, leading to the occurrence of seasonal influenza. However, fatal influenza is usually characterized by pneumonia involving alveoli, which can cause respiratory failure [92,105].
SARS-CoV-2
Respiratory tract and lungs: The lungs showed consolidation in varying degrees. With the naked eye, the lung can be viewed with gray-white lesions and dark red bleeding [113]. Ackermann’s study found that the average weight of COVID-19 lungs was significantly higher than that of healthy people and lower than that of patients with influenza pneumonia [114].
The early pathological manifestations of COVID-19 were pulmonary edema, obvious protein exudation, vascular hyperemia, focal pneumocytes reactive proliferation, and patchy inflammatory cell infiltration, accompanied by inflammatory cluster fibrin-like substance and multinucleated giant cell formation [115]. With the development of the disease, the lungs may show DAD and perivascular lymphocyte infiltration. Moreover, there were more ACE2 positive lymphocytes in alveolar epithelial cells and capillary endothelial cells in COVID-19 and influenza pneumonia than in healthy people [114]. As for angiocentric inflammation and thrombosis, COVID-19 presents unique features of them, such as severe vascular endothelial injury and endothelial cell membrane destruction related to intracellular SARS-CoV-2 virus. Meanwhile, like influenza pneumonia, COVID-19 can present extensive fibrin thrombosis and thrombosis, accompanied by microvascular lesions and alveolar-capillary occlusion in the lungs. The incidence of microthrombus in alveolar capillaries was 9 times higher than that in influenza, while the number of thrombus in venule behind pulmonary capillaries less than a diameter of 1 mm was significantly lower than that in influenza. Specifically, the lungs of COVID-19 can not only generate neovascularization through conventional sprouting but also through intussusceptive angiogenesis, both of which are stronger than influenza. And the higher degree of pulmonary endothelialitis and thrombosis in patients with COVID-19 may be related to the observed relative frequency of sprouting and intussusceptive angiogenesis [114]. Consistent with the above, many studies showed that COVID-19 patients have coagulopathy, and COVID-19-related coagulopathy is a combination of mild diffuse intravascular coagulation(DIC) and localized pulmonary thrombotic microangiopathy, which may cause organ dysfunction [116,117].
Other organs
Except for pulmonary lesions, SARS-COV-2 also involves many organs such as immune organs, cardiovascular system, liver, and kidney. The pathology of the spleen showed a significant decrease in the number of lymphocytes, degeneration, and necrosis. The number of three-line cells in bone marrow can reduce to different degrees [118]. bAs for the heart, cardiomyocytes can be deformed and necrotic, and the cardiovascular system can show chronic damage [119]. Xu et al found that SARS-COV-2 may cause liver injury through direct cytopathic effect and/or immunopathological effect caused by excessive inflammatory response [120]. Therefore, hepatocyte degeneration and focal necrosis can be seen in the liver, and bile thrombus can be seen in the bile duct. There transparent thrombus in the glomerular capillaries and protein exudate presents in the renal glomerular sac lumen. And renal tubular epithelial can degenerate and exfoliate. Importantly, SARS-CoV-2 was not detected in all of the above issues. The mucosal epithelium of the esophagus, stomach, and intestine degenerated, necrotic, and exfoliated to different degrees [17,18,118].
Briefly, endothelial injury, micro-angiopathy, angiogenesis, and coagulopathy help to distinguish the lung pathology of COVID-19 from that of equal severe influenza, and the pathological manifestations of COVID-19 are more serious than those of influenza, the severity is more similar to avian influenza, which can involve deep airway and alveoli. This explains why avian influenza and COVID-19 can present severe pneumonia, easily causing significant hypoxemia and respiratory failure [121,122] (Table 3).
Clinical features and treatment
Influenza: The clinical manifestations of influenza are characterized by various systemic symptoms, such as fever, chills, and a series of respiratory symptoms. However, since influenza viruses usually only affect larger airways, the infection is mild. Patients with A/H1N1 influenza usually show influenza-like symptoms and may have complications such as pneumonia. A few patients progress rapidly, underlying chronic lung disease worsens, lung function decreases, hypoxemia, acute lung injury (ALI)/ARDS, respiratory failure, MODS, and so on [4,82,123,124]. Moreover, non-respiratory complications are rare but can be devastating, which result from the influenza virus or invasive bacteria while compromised with influenza [125]. And life-threatening H1N1 influenza can manifest characteristics of macrophage activation syndrome(MAS)/haemophagocytic lymphohistocytosis(HLH), including hyper inflammation, pancytopenia, coagulopathy, and liver dysfunction, but the frequency and pathologic basis of these findings remain undefined [126]. Of note, avian influenza also presents with fever and respiratory symptoms, but it can worse rapidly, and carries a high risk of death [121,127]. For the influenza epidemic, routine vaccination is recommended [125]. Most people who are otherwise healthy and have the flu do not need antiviral drugs. Currently, the CDC recommends four FDA-approved antiviral drugs, including oseltamivir phosphate, zanamivir, peramivir, and balosavir marboxil, and recommends timely treatment for influenza-infected or suspected influenza-infected people, and who are at high risk of serious flu complications, such as people with asthma, diabetes or heart disease [10].
SARS-CoV-2
The most common symptoms of COVID-19 were fever, cough, fatigue, and myalgia [60,100]. In fact, COVID-19 initially presents with “flu-like” symptoms and then progresses to life-threatening systemic inflammation and multiple organ dysfunction [128]. It is worth noting that SARS-CoV-2 infection can show five different phenotypes, the most common and benign phenotype is phenotype 1, mainly characterized by fever, headache, or mild respiratory symptoms and no hypoxemia. Phenotype 2, which accounts for 80% of inpatients, is characterized by hypoxemia or small opacities on chest X-ray, while phenotype 3 presents more severe hypoxemia and higher respiratory frequency. Phenotype 4 is characterized by severe hypoxemia requiring mechanical ventilation. Phenotype 5 is the late stage of ALI. Thus, each phenotype should have a corresponding treatment scheme, which will be more helpful for clinicians and researchers to optimize the treatment of patients and improve their prognosis [129]. In addition, Gattinoni et al. divide covid-19 pneumonia into Type L featured by Low Elastance, Low ventilation-to-perfusion ratio, Low lung weight, and Low recruit ability, and Type H featured by High elastance, High right-to-left shunt, High lung weight and High recruitability [130]. However, this classification needs to be studied [131]. As for complications, it was mentioned earlier that patients with COVID-19 have coagulation abnormalities and a high incidence of thromboembolism [132]. Accordingly, Tang et al found that coagulation abnormalities, especially markedly elevated D-dimer and fibrin degradation product(FDP) are common in deaths with COVID-19 [133]. And the level of D-dimer is a good index for predicting venous thromboembolism(VTE) in patients with severe COVID-19 [134], showing that severe COVID-19 is often complicated with coagulopathy and abnormal coagulation parameters are associated with poor prognosis [133]. Besides, severe COVID-19-associated pneumonia may exhibit features of systemic hyper-inflammation designated under the umbrella term of MAS or “cytokine storm”, also known as secondary HLH [135]. COVID-19 mortality may be due to virus-activated cytokine storm or fulminant myocarditis [136]. It can be seen that for treatment, in addition to recommending the use of monoclonal antibodies against spike protein for immune interventions, we can also target CRS cytokines for immunotherapy. So far, it has been found that immunoglobulin and plasma therapy can improve the prognosis of COVID-19 patients, while scientific and systemic antiviral strategies are still being studied [137-139].
Influenza occurs almost every season, and many people around the world have established immunity to seasonal influenza strains. And there are mature vaccines to help the body develop immunity to influenza, and good progress has been made in antiviral therapy [28]. Convalescent plasma has also been proved to be useful in the treatment [140-142]. As for COVID-19, although a lot of clinical trials on various antiviral drugs, traditional Chinese medicine and vaccines have been launched, and some may have satisfactory results [143-147]. As of May 27, 2021, China has administered more than 3.3 billion doses of the novel coronavirus vaccine [148]. However, studies have shown that even vaccinated people can become infected with new coronavirus variants, so further research is needed on specific inhibitors of SARS-CoV-2 [103] (Table 3). With regard to the prognosis of COVID-19, studies have shown that age and male are independent risk factors for death [149]. A study also claimed that most of the critically ill patients were elderly men, and the mortality rate in intensive care units requiring mechanical ventilation and high levels of PEEP was 26% [150]. In addition, the presence of various SARS-CoV-2 strains and immune system dysfunction leading to the possibility of a recurrence of COVID-19. Combined with the experience of fighting SARS and MERS, for example, discharged SARS patients should still be isolated at home, and MERS patients should be discharged with strict viral nucleic acid testing procedures [151]. Therefore, the control of discharge criteria should be cautious, it is not recommended that COVID-19 patients who meet the discharge criteria discharge immediately [152,153], and discharging should not be regarded as the end point of fighting the virus. Reasonable and scientific long-term follow-up strategies are necessary for coronavirus control [151].
Conclusion
As is known, influenza occurs every year, and influenza pandemics occur irregularly. It is precise because of this that we know more about influenza viruses. Unlike highly virulent viruses, uncomplicated human influenza viruses mainly affect larger airways and rarely affect alveoli. However, SARS-CoV-2 mainly involves deep airways and lungs, which can cause DAD, resulting in severe hypoxemia. It can be seen that SASR-CoV-2 has more severe damage to lung function, and a higher rate of disability, mortality, and severe illness.
From the six aspects of transmission, pathogenesis, laboratory tests, imaging tests, pathological findings, and clinical features and treatment, it can be inferred that SARS-CoV-2 is no less contagious than the influenza virus. Moreover, the destructive power of SARS-CoV-2 is no less than that of the avian influenza virus. What is worse, because of the emergence of SARS-CoV-2 mutants, vaccination cannot resist the attack of the virus[12]. Moreover, according to epidemiological data search and literature review, there is no specific inhibitor against SARS-CoV-2. Due to this, we could realize that we are not as good at dealing with SARS-CoV-2 as we are with influenza virus and the top priority is to prevent the further spread of SASA-CoV-2. Meanwhile, the WHO is constantly updating the latest prevention and control strategies, emphasizing that prevention is more important than treatment at this stage. To prevent and control emerging infectious diseases COVID-19, the most classic method (three principles) is the most effective. Firstly, control the source of infection. Such as rapid identification of cases, rapid, effective, and strict detection and isolation of confirmed and suspected patients, and comprehensive identification, tracking, and quarantine of contacts. Secondly, cut off the routes of transmission. For example, work suspension, school suspension and traffic restrictions, attention to hand hygiene, and respiratory hygiene etiquette. Thirdly, protect the susceptible. As the crowd is generally susceptible to SARS-CoV-2, governments restrict the unnecessary travel of residents, advise people to keep a certain distance from each other, and recommend people improve their own immunity, so as to protect the susceptible population [154]. So far, many countries have gradually taken relevant preventive measures to protect the public. From the point of view of epidemic control, we should recognize that it is necessary to take early action and implement comprehensive public health measures. As for the “herd immunity” policy, it is difficult to implement because general immunity is difficult to obtain, and the vaccines currently used in clinical work are difficult to help establish effective immunity due to the continuous emergence of virus variants.
Currently, the epidemic is still severe. The epidemic prevention war is continuing. Each country is at different stages of the outbreak. The anti-epidemic experience of various countries and the strategies published by WHO are still constantly updated. Real-time tracking of research and epidemiology will help us learn more about SARS-CoV-2. Although many treatments and experiences have been proposed, the only measure that is feasible at present seems to be strict isolation of the general population. Although many treatments and experiences have been proposed, the confirmed feasible measure seems to be strict quarantine measures for the general population. However, as containment measures were achieved relatively successfully, we at least could recognize that controlling the source of infection, cutting off the routes of transmission, and protecting susceptible populations is the key to fighting SARS-CoV-2. At the same time, we should continue to focus on vaccine development and research of antiviral drugs.
Support statement: This work was supported by the National Natural Science Foundation of China (81970015).
- World Health Organization, Coronavirus disease (COVID-19) Pandemic.
- World Health Organization, Coronavirus disease (COVID-19) Pandemic.
- Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, et al. Clinical Characteristics of Coronavirus Disease 2019 in China. The New England Journal of Medicine 2020.
- Cox NJ, Subbarao K. Influenza. Lancet. 1999 Oct 9;354(9186):1277-82. doi: 10.1016/S0140-6736(99)01241-6. PMID: 10520648.
- Killingley B, Nguyen-Van-Tam J. Routes of influenza transmission. Influenza Other Respir Viruses. 2013 Sep;7 Suppl 2(Suppl 2):42-51. doi: 10.1111/irv.12080. PMID: 24034483; PMCID: PMC5909391.
- Shinya K, Ebina M, Yamada S, Ono M, Kasai N et al. Influenza virus receptors in the human airway. Nature. 2006; 440: 435-436.
- Sullivan KM, Monto AS, Longini IM Jr. Estimates of the US health impact of influenza. Am J Public Health. 1993 Dec;83(12):1712-6. doi: 10.2105/ajph.83.12.1712. PMID: 8259800; PMCID: PMC1694912.
- Tokars JI, Olsen SJ, Reed C. Seasonal Incidence of Symptomatic Influenza in the United States. Clin Infect Dis. 2018 May 2;66(10):1511-1518. doi: 10.1093/cid/cix1060. PMID: 29206909; PMCID: PMC5934309.
- Centers for Disease Control and Prevention (CDC). Estimates of deaths associated with seasonal influenza --- United States, 1976-2007. MMWR Morb Mortal Wkly Rep. 2010 Aug 27;59(33):1057-62. PMID: 20798667.
- Centres for Disease Control and Prevention (CDC), Influenza (Flu).
- Erçen Diken Ö, Arslan S, Akdoğan Ö, Yapar D, Ünal Ö, Demir E, Baykam N. Clinical, radiological and prognostic features of influenza cases in the influenza epidemic during years 2016-2017. Tuberk Toraks. 2018 Jun;66(2):144-149. English. doi: 10.5578/tt.66122. PMID: 30246658.
- Araf Y, Akter F, Tang YD, Fatemi R, Parvez MSA, Zheng C, Hossain MG. Omicron variant of SARS-CoV-2: Genomics, transmissibility, and responses to current COVID-19 vaccines. J Med Virol. 2022 May;94(5):1825-1832. doi: 10.1002/jmv.27588. Epub 2022 Jan 23. PMID: 35023191; PMCID: PMC9015557.
- Carlos WG, Dela Cruz CS, Cao B, Pasnick S, Jamil S. Novel Wuhan (2019-nCoV) Coronavirus. Am J Respir Crit Care Med. 2020 Feb 15;201(4):P7-P8. doi: 10.1164/rccm.2014P7. PMID: 32004066.
- Wax RS, Christian MD. Practical recommendations for critical care and anesthesiology teams caring for novel coronavirus (2019-nCoV) patients. Can J Anaesth. 2020 May;67(5):568-576. doi: 10.1007/s12630-020-01591-x. Epub 2020 Feb 12. PMID: 32052373; PMCID: PMC7091420.
- Novel coronavirus may spread via digestive system: experts.
- Liu Y, Ning Z, Chen Y, Guo M, Liu Y, Gali NK, Sun L, Duan Y, Cai J, Westerdahl D, Liu X, Xu K, Ho KF, Kan H, Fu Q, Lan K. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature. 2020 Jun;582(7813):557-560. doi: 10.1038/s41586-020-2271-3. Epub 2020 Apr 27. PMID: 32340022.
- Diao B, Wang C, Wang R, Feng Z, Zhang J, Yang H, Tan Y, Wang H, Wang C, Liu L, Liu Y, Liu Y, Wang G, Yuan Z, Hou X, Ren L, Wu Y, Chen Y. Human kidney is a target for novel severe acute respiratory syndrome coronavirus 2 infection. Nat Commun. 2021 May 4;12(1):2506. doi: 10.1038/s41467-021-22781-1. PMID: 33947851; PMCID: PMC8096808.
- National Health Commission of the People’s Republic of China. Diagnosis and treatment protocols of pneumonia caused by SARS-CoV-2 (trial version 8 ).
- World Health Organization, Report of the WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19).
- Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, Xing X, Xiang N, Wu Y, Li C, Chen Q, Li D, Liu T, Zhao J, Liu M, Tu W, Chen C, Jin L, Yang R, Wang Q, Zhou S, Wang R, Liu H, Luo Y, Liu Y, Shao G, Li H, Tao Z, Yang Y, Deng Z, Liu B, Ma Z, Zhang Y, Shi G, Lam TTY, Wu JT, Gao GF, Cowling BJ, Yang B, Leung GM, Feng Z. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med. 2020 Mar 26;382(13):1199-1207. doi: 10.1056/NEJMoa2001316. Epub 2020 Jan 29. PMID: 31995857; PMCID: PMC7121484.
- Prompetchara E, Ketloy C, Palaga T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol. 2020 Mar;38(1):1-9. doi: 10.12932/AP-200220-0772. PMID: 32105090.
- Li YK, Peng S, Li LQ, Wang Q, Ping W, Zhang N, Fu XN. Clinical and Transmission Characteristics of Covid-19 - A Retrospective Study of 25 Cases from a Single Thoracic Surgery Department. Curr Med Sci. 2020 Apr;40(2):295-300. doi: 10.1007/s11596-020-2176-2. Epub 2020 Mar 30. PMID: 32232652; PMCID: PMC7104422.
- Ndaïrou F, Area I, Nieto JJ, Torres DFM. Mathematical modeling of COVID-19 transmission dynamics with a case study of Wuhan. Chaos Solitons Fractals. 2020 Jun;135:109846. doi: 10.1016/j.chaos.2020.109846. Epub 2020 Apr 27. PMID: 32341628; PMCID: PMC7184012.
- Al-Tawfiq JA, Rodriguez-Morales AJ. Super-spreading events and contribution to transmission of MERS, SARS, and SARS-CoV-2 (COVID-19). J Hosp Infect. 2020 Jun;105(2):111-112. doi: 10.1016/j.jhin.2020.04.002. Epub 2020 Apr 8. PMID: 32277963; PMCID: PMC7194732.
- Biggerstaff M, Cauchemez S, Reed C, Gambhir M, Finelli L. Estimates of the reproduction number for seasonal, pandemic, and zoonotic influenza: a systematic review of the literature. BMC Infect Dis. 2014 Sep 4;14:480. doi: 10.1186/1471-2334-14-480. PMID: 25186370; PMCID: PMC4169819.
- Taubenberger JK, Morens DM. The 1918 Influenza Pandemic and Its Legacy. Cold Spring Harb Perspect Med. 2020 Oct 1;10(10):a038695. doi: 10.1101/cshperspect.a038695. PMID: 31871232; PMCID: PMC7528857.
- Hsu LY, Chia PY, Lim JF. The Novel Coronavirus (SARS-CoV-2) Epidemic. Ann Acad Med Singap. 2020 Mar 16;49(3):105-107. PMID: 32200398.
- Zambon MC. Epidemiology and pathogenesis of influenza. J Antimicrob Chemother. 1999 Nov;44 Suppl B:3-9. doi: 10.1093/jac/44.suppl_2.3. PMID: 10877456.
- Rogers GN, Paulson JC. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology. 1983 Jun;127(2):361-73. doi: 10.1016/0042-6822(83)90150-2. PMID: 6868370.
- Zambon MC. The pathogenesis of influenza in humans. Rev Med Virol. 2001 Jul-Aug;11(4):227-41. doi: 10.1002/rmv.319. PMID: 11479929.
- Aegerter H, Kulikauskaite J, Crotta S, Patel H, Kelly G, Hessel EM, Mack M, Beinke S, Wack A. Influenza-induced monocyte-derived alveolar macrophages confer prolonged antibacterial protection. Nat Immunol. 2020 Feb;21(2):145-157. doi: 10.1038/s41590-019-0568-x. Epub 2020 Jan 13. PMID: 31932810; PMCID: PMC6983324.
- Shi C, Pamer EG. Monocyte recruitment during infection and inflammation. Nat Rev Immunol. 2011 Oct 10;11(11):762-74. doi: 10.1038/nri3070. PMID: 21984070; PMCID: PMC3947780.
- Lin KL, Suzuki Y, Nakano H, Ramsburg E, Gunn MD. CCR2+ monocyte-derived dendritic cells and exudate macrophages produce influenza-induced pulmonary immune pathology and mortality. J Immunol. 2008 Feb 15;180(4):2562-72. doi: 10.4049/jimmunol.180.4.2562. PMID: 18250467.
- Hoeve MA, Nash AA, Jackson D, Randall RE, Dransfield I. Influenza virus A infection of human monocyte and macrophage subpopulations reveals increased susceptibility associated with cell differentiation. PLoS One. 2012;7(1):e29443. doi: 10.1371/journal.pone.0029443. Epub 2012 Jan 4. PMID: 22238612; PMCID: PMC3251590.
- Lee LY, Ha do LA, Simmons C, de Jong MD, Chau NV, Schumacher R, Peng YC, McMichael AJ, Farrar JJ, Smith GL, Townsend AR, Askonas BA, Rowland-Jones S, Dong T. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J Clin Invest. 2008 Oct;118(10):3478-90. doi: 10.1172/JCI32460. Erratum in: J Clin Invest. 2012 Nov 1;122(11):4301. PMID: 18802496; PMCID: PMC2542885.
- Jansen JM, Gerlach T, Elbahesh H, Rimmelzwaan GF, Saletti G. Influenza virus-specific CD4+ and CD8+ T cell-mediated immunity induced by infection and vaccination. J Clin Virol. 2019 Oct;119:44-52. doi: 10.1016/j.jcv.2019.08.009. Epub 2019 Aug 24. PMID: 31491709.
- Wilkinson TM, Li CK, Chui CS, Huang AK, Perkins M, Liebner JC, Lambkin-Williams R, Gilbert A, Oxford J, Nicholas B, Staples KJ, Dong T, Douek DC, McMichael AJ, Xu XN. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med. 2012 Jan 29;18(2):274-80. doi: 10.1038/nm.2612. PMID: 22286307.
- Juno JA, van Bockel D, Kent SJ, Kelleher AD, Zaunders JJ, Munier CM. Cytotoxic CD4 T Cells-Friend or Foe during Viral Infection? Front Immunol. 2017 Jan 23;8:19. doi: 10.3389/fimmu.2017.00019. PMID: 28167943; PMCID: PMC5253382.
- Brown DM, Lampe AT, Workman AM. The Differentiation and Protective Function of Cytolytic CD4 T Cells in Influenza Infection. Front Immunol. 2016 Mar 9;7:93. doi: 10.3389/fimmu.2016.00093. PMID: 27014272; PMCID: PMC4783394.
- Brown DM, Dilzer AM, Meents DL, Swain SL. CD4 T cell-mediated protection from lethal influenza: perforin and antibody-mediated mechanisms give a one-two punch. J Immunol. 2006: 177:2888-2898.
- Brown DM, Kamperschroer C, Dilzer AM, Roberts DM, Swain SL. IL-2 and antigen dose differentially regulate perforin- and FasL-mediated cytolytic activity in antigen specific CD4+ T cells. Cell Immunol. 2009;257(1-2):69-79. doi: 10.1016/j.cellimm.2009.03.002. Epub 2009 Mar 31. PMID: 19338979; PMCID: PMC2683476.
- Sun JC, Bevan MJ. Defective CD8 T cell memory following acute infection without CD4 T cell help. Science. 2003 Apr 11;300(5617):339-42. doi: 10.1126/science.1083317. PMID: 12690202; PMCID: PMC2778341.
- Shedlock DJ, Shen H. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science. 2003 Apr 11;300(5617):337-9. doi: 10.1126/science.1082305. PMID: 12690201.
- Riberdy JM, Christensen JP, Branum K, Doherty PC. Diminished primary and secondary influenza virus-specific CD8(+) T-cell responses in CD4-depleted Ig(-/-) mice. J Virol. 2000 Oct;74(20):9762-5. doi: 10.1128/jvi.74.20.9762-9765.2000. PMID: 11000251; PMCID: PMC112411.
- Belz GT, Wodarz D, Diaz G, Nowak MA, Doherty PC. Compromised influenza virus-specific CD8(+)-T-cell memory in CD4(+)-T-cell-deficient mice. J Virol 2002, 76:12388-12393.
- Devarajan P, Jones MC, Kugler-Umana O, Vong AM, Xia J, Swain SL. Pathogen Recognition by CD4 Effectors Drives Key Effector and Most Memory Cell Generation Against Respiratory Virus. Front Immunol. 2018 Mar 26;9:596. doi: 10.3389/fimmu.2018.00596. PMID: 29632538; PMCID: PMC5879149.
- Yin L, Zheng D, Limmon GV, Leung NH, Xu S, Rajapakse JC, Yu H, Chow VT, Chen J. Aging exacerbates damage and delays repair of alveolar epithelia following influenza viral pneumonia. Respir Res. 2014 Sep 30;15(1):116. doi: 10.1186/s12931-014-0116-z. PMID: 25265939; PMCID: PMC4189598.
- Li G, Fan Y, Lai Y, Han T, Li Z, Zhou P, Pan P, Wang W, Hu D, Liu X, Zhang Q, Wu J. Coronavirus infections and immune responses. J Med Virol. 2020 Apr;92(4):424-432. doi: 10.1002/jmv.25685. Epub 2020 Feb 7. PMID: 31981224; PMCID: PMC7166547.
- Chen Y, Guo Y, Pan Y, Zhao ZJ. Structure analysis of the receptor binding of 2019-nCoV. Biochem Biophys Res Commun. 2020 Feb 17;525(1):135–40. doi: 10.1016/j.bbrc.2020.02.071. Epub ahead of print. PMID: 32081428; PMCID: PMC7092824.
- Yang N, Shen HM. Targeting the Endocytic Pathway and Autophagy Process as a Novel Therapeutic Strategy in COVID-19. Int J Biol Sci. 2020 Mar 15;16(10):1724-1731. doi: 10.7150/ijbs.45498. PMID: 32226290; PMCID: PMC7098027.
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 Mar;579(7798):270-273. doi: 10.1038/s41586-020-2012-7. Epub 2020 Feb 3. PMID: 32015507; PMCID: PMC7095418.
- Zou X, Chen K, Zou J, Han P, Hao J, Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med. 2020 Apr;14(2):185-192. doi: 10.1007/s11684-020-0754-0. Epub 2020 Mar 12. PMID: 32170560; PMCID: PMC7088738.
- Zhang H, Kang Z, Gong H, Xu D, Wang J et al. The digestive system is a potential route of 2019-nCov infection: a bioinformatics analysis based on single-cell transcriptomes. bioRxiv. 2020:2020.2001.2030.927806.
- Zhao Y, Zhao Z, Wang Y, Zhou Y, Ma Y, et al. Single-cell RNA expression profiling of ACE2, the putative receptor of Wuhan 2019-nCov. bioRxiv. 2020:2020.2001.2026.919985.
- Chai X, Hu L, Zhang Y, Han W, Lu Z et al. Specific ACE2 Expression in Cholangiocytes May Cause Liver Damage After 2019-nCoV Infection. bioRxiv 2020:2020.2002.2003.931766.
- Zheng M, Gao Y, Wang G, Song G, Liu S, Sun D, Xu Y, Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol. 2020 May;17(5):533-535. doi: 10.1038/s41423-020-0402-2. Epub 2020 Mar 19. PMID: 32203188; PMCID: PMC7091858.
- Romagnani S. Regulation of the T cell response. Clin Exp Allergy. 2006 Nov;36(11):1357-66. doi: 10.1111/j.1365-2222.2006.02606.x. PMID: 17083345.
- Liu B, Li M, Zhou Z, Guan X, Xiang Y. Can we use interleukin-6 (IL-6) blockade for coronavirus disease 2019 (COVID-19)-induced cytokine release syndrome (CRS)? J Autoimmun. 2020 Jul;111:102452. doi: 10.1016/j.jaut.2020.102452. Epub 2020 Apr 10. PMID: 32291137; PMCID: PMC7151347.
- Channappanavar R, Fehr AR, Vijay R, Mack M, Zhao J, Meyerholz DK, Perlman S. Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe. 2016 Feb 10;19(2):181-93. doi: 10.1016/j.chom.2016.01.007. PMID: 26867177; PMCID: PMC4752723.
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 Feb 15;395(10223):497-506. doi: 10.1016/S0140-6736(20)30183-5. Epub 2020 Jan 24. Erratum in: Lancet. 2020 Jan 30;: PMID: 31986264; PMCID: PMC7159299.
- From the autopsy of SARS, a brief analysis of coronavirus disease-19 (COVID-19).
- Srikiatkhachorn A, Mathew A, Rothman AL. Immune-mediated cytokine storm and its role in severe dengue. Semin Immunopathol. 2017 Jul;39(5):563-574. doi: 10.1007/s00281-017-0625-1. Epub 2017 Apr 11. PMID: 28401256; PMCID: PMC5496927.
- Chousterman BG, Swirski FK, Weber GF. Cytokine storm and sepsis disease pathogenesis. Semin Immunopathol. 2017 Jul;39(5):517-528. doi: 10.1007/s00281-017-0639-8. Epub 2017 May 29. PMID: 28555385.
- Fu Y, Cheng Y, Wu Y. Understanding SARS-CoV-2-Mediated Inflammatory Responses: From Mechanisms to Potential Therapeutic Tools. Virol Sin. 2020 Jun;35(3):266-271. doi: 10.1007/s12250-020-00207-4. Epub 2020 Mar 3. PMID: 32125642; PMCID: PMC7090474.
- Liu L, Wei Q, Lin Q, Fang J, Wang H, Kwok H, Tang H, Nishiura K, Peng J, Tan Z, Wu T, Cheung KW, Chan KH, Alvarez X, Qin C, Lackner A, Perlman S, Yuen KY, Chen Z. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019 Feb 21;4(4):e123158. doi: 10.1172/jci.insight.123158. PMID: 30830861; PMCID: PMC6478436.
- van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, Kuiken T. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am J Pathol. 2007 Oct;171(4):1215-23. doi: 10.2353/ajpath.2007.070248. Epub 2007 Aug 23. PMID: 17717141; PMCID: PMC1988871.
- Yun TJ, Park CM, Kwon GJ, Woo SK, Park SH, Choi SH, Lee HJ, Goo JM. Clinical and radiological features of pandemic H1N1 2009 influenza virus infection manifesting as acute febrile respiratory illness at their initial presentations: comparison with contemporaneous non-H1N1 patients. Acta Radiol. 2011 May 1;52(4):410-6. doi: 10.1258/ar.2011.100411. Epub 2011 Mar 9. PMID: 21498293.
- Gao HN, Lu HZ, Cao B, Du B, Shang H, Gan JH, Lu SH, Yang YD, Fang Q, Shen YZ, Xi XM, Gu Q, Zhou XM, Qu HP, Yan Z, Li FM, Zhao W, Gao ZC, Wang GF, Ruan LX, Wang WH, Ye J, Cao HF, Li XW, Zhang WH, Fang XC, He J, Liang WF, Xie J, Zeng M, Wu XZ, Li J, Xia Q, Jin ZC, Chen Q, Tang C, Zhang ZY, Hou BM, Feng ZX, Sheng JF, Zhong NS, Li LJ. Clinical findings in 111 cases of influenza A (H7N9) virus infection. N Engl J Med. 2013 Jun 13;368(24):2277-85. doi: 10.1056/NEJMoa1305584. Epub 2013 May 22. Erratum in: N Engl J Med. 2013 Nov 7;369(19):1869. PMID: 23697469.
- Durani U, Dioverti Prono MV, Tosh PK, Patnaik M, Barreto JN, Tande AJ. Influenza infection in neutropenic adults. Infect Dis (Lond). 2017 Feb;49(2):141-146. doi: 10.1080/23744235.2016.1231418. Epub 2016 Sep 16. PMID: 27636702.
- Cheng Y, Zhao H, Song P, Zhang Z, Chen J, Zhou YH. Dynamic changes of lymphocyte counts in adult patients with severe pandemic H1N1 influenza A. J Infect Public Health. 2019 Nov-Dec;12(6):878-883. doi: 10.1016/j.jiph.2019.05.017. Epub 2019 Jun 13. PMID: 31202719; PMCID: PMC7102863.
- Gonzalez Y, Juárez E, Carranza C, Sada E, Pedraza-Sánchez S, Torres M. Diminished effector and memory CD8+ circulating T lymphocytes in patients with severe influenza caused by the AH1N1 pdm09 virus. Virology. 2017 Jan;500:139-148. doi: 10.1016/j.virol.2016.10.016. Epub 2016 Nov 2. PMID: 27816639.
- Peaper DR, Landry ML. Rapid Diagnosis of Influenza. Clinics in Laboratory Medicine. 34: 365-385.
- Playford EG, Dwyer DE. Laboratory diagnosis of influenza virus infection. Pathology. 2002 Apr;34(2):115-25. doi: 10.1080/003130201201117909. PMID: 12009091.
- Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020 Feb 15;395(10223):507-513. doi: 10.1016/S0140-6736(20)30211-7. Epub 2020 Jan 30. PMID: 32007143; PMCID: PMC7135076.
- Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, Zhao Y, Li Y, Wang X, Peng Z. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 2020 Mar 17;323(11):1061-1069. doi: 10.1001/jama.2020.1585. Erratum in: JAMA. 2021 Mar 16;325(11):1113. PMID: 32031570; PMCID: PMC7042881.
- Wan Q, Shi A, He T, Tang L. Clinical features of 153 patients with COVID-19 in Chongqing municipality. Chin J Clin Infect Dis. 2020; 13 (2020-02-28).
- Zhou S, Wang Y, Zhu T, Xia L. CT Features of Coronavirus Disease 2019 (COVID-19) Pneumonia in 62 Patients in Wuhan, China. AJR Am J Roentgenol. 2020 Jun;214(6):1287-1294. doi: 10.2214/AJR.20.22975. Epub 2020 Mar 5. PMID: 32134681.
- Zhou R, To KK, Wong YC, Liu L, Zhou B et al. Acute SARS-CoV-2 Infection Impairs Dendritic Cell and T Cell Responses. Immunity. 2020; 53:864-877.e865.
- Wang F, Nie J, Wang H, Zhao Q, Xiong Y, Deng L, Song S, Ma Z, Mo P, Zhang Y. Characteristics of Peripheral Lymphocyte Subset Alteration in COVID-19 Pneumonia. J Infect Dis. 2020 May 11;221(11):1762-1769. doi: 10.1093/infdis/jiaa150. PMID: 32227123; PMCID: PMC7184346.
- Cirit OS, Mutlu E, Sancak B, Kocagöz T, Can Ö, Çicek C, Arzu Sayiner A, Appak Ö, Uyar NY, Külah C, Çiçek AÇ, Özgümüs OB, Ay Altintop Y, Saatçi E, Karsligil T, Zer Y, Özen NS, Çekin Y, Karahan ZC, Evren E, Karakoç AE, Orhan SG, Mutlu D, Bozdemir T, Çayci YT, Çinar C, Tasbakan M, Mert M, Çinar E, Kutsoylu OÖE, Kocagöz S, Ertürk A, Çelik I, Mete AÖ, Günalp Eneyli M, Akdemir I, Karakök T, Inan D, Atilla A, Taflan ŞO, Yörük KE. Comparison of a novel antigen detection test with reverse transcription polymerase chain reaction assay for laboratory diagnosis of SARS-CoV-2 infection. Infection. 2022 May 5:1–6. doi: 10.1007/s15010-022-01832-9. Epub ahead of print. PMID: 35513690; PMCID: PMC9070611.
- Tan DL, Wang F, Liu ZG, Tao FX. Application and suggestion of standard chest imaging examination in diagnosis and treatment of swine influenza H1N1. Chinese Journal of Medical Imaging Technology. 2015; 31(04):563-566.
- Lu PX, Luo YT, Zheng QT. Image manifestations of Influenza and the key points of the latest National Diagnostic and Therapeutic schemes. Electronic Journal of Emerging Infectious Diseases. 2019; 4(1): 56-61.
- Koo HJ, Lim S, Choe J, Choi SH, Sung H, Do KH. Radiographic and CT Features of Viral Pneumonia. Radiographics. 2018 May-Jun;38(3):719-739. doi: 10.1148/rg.2018170048. PMID: 29757717.
- Kang H, Lee KS, Jeong YJ, Lee HY, Kim KI, Nam KJ. Computed tomography findings of influenza A (H1N1) pneumonia in adults: pattern analysis and prognostic comparisons. J Comput Assist Tomogr. 2012 May-Jun;36(3):285-90. doi: 10.1097/RCT.0b013e31825588e6. PMID: 22592609.
- Oliveira EC, Marik PE, Colice G. Influenza pneumonia: a descriptive study. Chest. 2001 Jun;119(6):1717-23. doi: 10.1378/chest.119.6.1717. PMID: 11399696.
- Cho WH, Kim YS, Jeon DS, Kim JE, Kim KI, Seol HY, Kim KU, Park HK, Lee MK, Park SK, Jeong YJ. Outcome of pandemic H1N1 pneumonia: clinical and radiological findings for severity assessment. Korean J Intern Med. 2011 Jun;26(2):160-7. doi: 10.3904/kjim.2011.26.2.160. Epub 2011 Jun 1. PMID: 21716592; PMCID: PMC3110848.
- Rohani P, Jude CM, Chan K, Barot N, Kamangar N. Chest Radiological Findings of Patients With Severe H1N1 Pneumonia Requiring Intensive Care. J Intensive Care Med. 2016 Jan;31(1):51-60. doi: 10.1177/0885066614538753. Epub 2014 Jun 12. PMID: 24923491.
- Qureshi NR, Hien TT, Farrar J, Gleeson FV. The radiologic manifestations of H5N1 avian influenza. J Thorac Imaging. 2006 Nov;21(4):259-64. doi: 10.1097/01.rti.0000213573.94032.53. PMID: 17110849.
- Xu SH, Li HJ, Li N, Hu CH, Li RT et al. Comparative study of CT findings and clinical course of patients with severe pneumonia due to avian influenza H7N9 and swine influenza H1N1 infection. Radiologic Practice. 2014; 29:(07):756-759.
- Lu PX, Zeng Z, Zheng FQ, Zheng GP, Zang J, et al. Characteristics of imaging manifestations and dynamic changes in patients with severe pneumonia caused by H7N9 avian influenza virus. Radiologic Practice, 2014; (07):740-744.
- Ajlan AM, Quiney B, Nicolaou S, Müller NL. Swine-origin influenza A (H1N1) viral infection: radiographic and CT findings. AJR Am J Roentgenol. 2009 Dec;193(6):1494-9. doi: 10.2214/AJR.09.3625. PMID: 19933639.
- Aviram G, Bar-Shai A, Sosna J, Rogowski O, Rosen G, Weinstein I, Steinvil A, Zimmerman O. H1N1 influenza: initial chest radiographic findings in helping predict patient outcome. Radiology. 2010 Apr;255(1):252-9. doi: 10.1148/radiol.10092240. PMID: 20308461.
- Song F, Shi N, Shan F, Zhang Z, Shen J, Lu H, Ling Y, Jiang Y, Shi Y. Emerging 2019 Novel Coronavirus (2019-nCoV) Pneumonia. Radiology. 2020 Apr;295(1):210-217. doi: 10.1148/radiol.2020200274. Epub 2020 Feb 6. Erratum in: Radiology. 2020 Dec;297(3):E346. PMID: 32027573; PMCID: PMC7233366.
- Wu J, Feng LC, Xian XY, Qiang J, Zhang J, Mao QX, Kong SF, Chen YC, Pan JP. [Novel coronavirus pneumonia (COVID-19) CT distribution and sign features]. Zhonghua Jie He He Hu Xi Za Zhi. 2020 Apr 12;43(4):321-326. Chinese. doi: 10.3760/cma.j.cn112147-20200217-00106. PMID: 32125131.
- Grillet F, Behr J, Calame P, Aubry S, Delabrousse E. Acute Pulmonary Embolism Associated with COVID-19 Pneumonia Detected with Pulmonary CT Angiography. Radiology. 2020 Sep;296(3):E186-E188. doi: 10.1148/radiol.2020201544. Epub 2020 Apr 23. PMID: 32324103; PMCID: PMC7233384.
- Marsico S, Espallargas Giménez I, Carbullanca Toledo SJ, Del Carpio Bellido LA, Maiques Llácer JM, Zuccarino F. Infarto pulmonar secundario a tromboembolia pulmonar en COVID-19 diagnosticada con angiotomografía computarizada pulmonar con energía dual [Pulmonary infarction secondary to pulmonary thromboembolism in COVID-19 diagnosed with dual-energy CT pulmonary angiography]. Rev Esp Cardiol. 2020 Aug;73(8):672-674. Spanish. doi: 10.1016/j.recesp.2020.04.011. Epub 2020 Jun 5. PMID: 32834362; PMCID: PMC7274630.
- Léonard-Lorant I, Delabranche X, Séverac F, Helms J, Pauzet C, Collange O, Schneider F, Labani A, Bilbault P, Molière S, Leyendecker P, Roy C, Ohana M. Acute Pulmonary Embolism in Patients with COVID-19 at CT Angiography and Relationship to d-Dimer Levels. Radiology. 2020 Sep;296(3):E189-E191. doi: 10.1148/radiol.2020201561. Epub 2020 Apr 23. PMID: 32324102; PMCID: PMC7233397.
- Tang GX, Li CH, Liu XY, Yang J, Shu WQ et al. Clinical and CT findings of coronavirus disease 2019. Chinese Journal of Respiratory and Critical Care Medicine. 2020; 19(02):161-165.
- Shen ZY, Yan XC, You XD, Zhang XW. CT Imaging Research Progress in COVID-19. Curr Med Imaging. 2022;18(3):267-274. doi: 10.2174/1573405617666210816091217. PMID: 34465280; PMCID: PMC8972255.
- Li K, Wu J, Wu F, Guo D, Chen L, Fang Z, Li C. The Clinical and Chest CT Features Associated With Severe and Critical COVID-19 Pneumonia. Invest Radiol. 2020 Jun;55(6):327-331. doi: 10.1097/RLI.0000000000000672. PMID: 32118615; PMCID: PMC7147273..
- van Wissen M, Keller TT, Ronkes B, Gerdes VE, Zaaijer HL, van Gorp EC, Brandjes DP, Levi M, Büller HR. Influenza infection and risk of acute pulmonary embolism. Thromb J. 2007 Oct 16;5:16. doi: 10.1186/1477-9560-5-16. PMID: 17939867; PMCID: PMC2104525.
- Cao B, Wang Y, Wen D, Liu W, Wang J et al. A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19. The New England Journal of Medicine. 2020; 382:1787-1799.
- Wen W, Chen C, Tang J, Wang C, Zhou M, Cheng Y, Zhou X, Wu Q, Zhang X, Feng Z, Wang M, Mao Q. Efficacy and safety of three new oral antiviral treatment (molnupiravir, fluvoxamine and Paxlovid) for COVID-19:a meta-analysis. Ann Med. 2022 Dec;54(1):516-523. doi: 10.1080/07853890.2022.2034936. PMID: 35118917; PMCID: PMC8820829.
- Kash JC, Taubenberger JK. The role of viral, host, and secondary bacterial factors in influenza pathogenesis. Am J Pathol. 2015 Jun;185(6):1528-36. doi: 10.1016/j.ajpath.2014.08.030. Epub 2015 Mar 5. PMID: 25747532; PMCID: PMC4450310.
- Taubenberger JK, Morens DM. The pathology of influenza virus infections. Annu Rev Pathol. 2008;3:499-522. doi: 10.1146/annurev.pathmechdis.3.121806.154316. PMID: 18039138; PMCID: PMC2504709.
- Nakajima N, Sato Y, Katano H, Hasegawa H, Kumasaka T, Hata S, Tanaka S, Amano T, Kasai T, Chong JM, Iizuka T, Nakazato I, Hino Y, Hamamatsu A, Horiguchi H, Tanaka T, Hasegawa A, Kanaya Y, Oku R, Oya T, Sata T. Histopathological and immunohistochemical findings of 20 autopsy cases with 2009 H1N1 virus infection. Mod Pathol. 2012 Jan;25(1):1-13. doi: 10.1038/modpathol.2011.125. Epub 2011 Aug 26. Erratum in: Mod Pathol. 2012 Mar;25(3):492. Iiduka, Toshihiko [corrected to Iizuka, Toshihiko]; Hasagawa, Akio [corrected to Hasegawa, Akio]. PMID: 21874012.
- Guarner J, Falcón-Escobedo R. Comparison of the pathology caused by H1N1, H5N1, and H3N2 influenza viruses. Arch Med Res. 2009; 40: 655-661.
- Franková V, Jirásek A, Tůmová B. Type A influenza: postmortem virus isolations from different organs in human lethal cases. Arch Virol. 1977;53(3):265-8. doi: 10.1007/BF01314671. PMID: 856111.
- Takahashi M, Yamada T, Nakashita Y, Saikusa H, Deguchi M, Kida H, Tashiro M, Toyoda T. Influenza virus-induced encephalopathy: clinicopathologic study of an autopsied case. Pediatr Int. 2000 Apr;42(2):204-14. doi: 10.1046/j.1442-200x.2000.01203.x. PMID: 10804743.
- Cioc AM, Nuovo GJ. Histologic and in situ viral findings in the myocardium in cases of sudden, unexpected death. Mod Pathol. 2002 Sep;15(9):914-22. doi: 10.1097/01.MP.0000024291.37651.CD. PMID: 12218208.
- Bowles NE, Ni J, Kearney DL, Pauschinger M, Schultheiss HP, McCarthy R, Hare J, Bricker JT, Bowles KR, Towbin JA. Detection of viruses in myocardial tissues by polymerase chain reaction. evidence of adenovirus as a common cause of myocarditis in children and adults. J Am Coll Cardiol. 2003 Aug 6;42(3):466-72. doi: 10.1016/s0735-1097(03)00648-x. PMID: 12906974.
- Kuiken T, Taubenberger JK. Pathology of human influenza revisited. Vaccine. 2008 Sep 12;26 Suppl 4(Suppl 4):D59-66. doi: 10.1016/j.vaccine.2008.07.025. PMID: 19230162; PMCID: PMC2605683.
- Liu Q, Wang RS, Qu GQ, Wang YY, Liu P et al. A general observation report on the systematic anatomy of COVID-19 's dead body. Journal of Forensic Medicine 2020; 36.
- Ackermann M, Verleden SE, Kuehnel M, Haverich A, Welte T, Laenger F, Vanstapel A, Werlein C, Stark H, Tzankov A, Li WW, Li VW, Mentzer SJ, Jonigk D. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in Covid-19. N Engl J Med. 2020 Jul 9;383(2):120-128. doi: 10.1056/NEJMoa2015432. Epub 2020 May 21. PMID: 32437596; PMCID: PMC7412750.
- Tian S, Hu W, Niu L, Liu H, Xu H, Xiao SY. Pulmonary Pathology of Early-Phase 2019 Novel Coronavirus (COVID-19) Pneumonia in Two Patients With Lung Cancer. J Thorac Oncol. 2020 May;15(5):700-704. doi: 10.1016/j.jtho.2020.02.010. Epub 2020 Feb 28. PMID: 32114094; PMCID: PMC7128866.
- Wichmann D, Sperhake JP, Lütgehetmann M, Steurer S, Edler C, Heinemann A, Heinrich F, Mushumba H, Kniep I, Schröder AS, Burdelski C, de Heer G, Nierhaus A, Frings D, Pfefferle S, Becker H, Bredereke-Wiedling H, de Weerth A, Paschen HR, Sheikhzadeh-Eggers S, Stang A, Schmiedel S, Bokemeyer C, Addo MM, Aepfelbacher M, Püschel K, Kluge S. Autopsy Findings and Venous Thromboembolism in Patients With COVID-19: A Prospective Cohort Study. Ann Intern Med. 2020 Aug 18;173(4):268-277. doi: 10.7326/M20-2003. Epub 2020 May 6. PMID: 32374815; PMCID: PMC7240772.
- Levi M, Thachil J, Iba T, Levy JH. Coagulation abnormalities and thrombosis in patients with COVID-19. Lancet Haematol. 2020 Jun;7(6):e438-e440. doi: 10.1016/S2352-3026(20)30145-9. Epub 2020 May 11. PMID: 32407672; PMCID: PMC7213964.
- Yao XH, Li TY, He ZC, Ping YF, Liu HW, Yu SC, Mou HM, Wang LH, Zhang HR, Fu WJ, Luo T, Liu F, Guo QN, Chen C, Xiao HL, Guo HT, Lin S, Xiang DF, Shi Y, Pan GQ, Li QR, Huang X, Cui Y, Liu XZ, Tang W, Pan PF, Huang XQ, Ding YQ, Bian XW. [A pathological report of three COVID-19 cases by minimal invasive autopsies]. Zhonghua Bing Li Xue Za Zhi. 2020 May 8;49(5):411-417. Chinese. doi: 10.3760/cma.j.cn112151-20200312-00193. PMID: 32172546.
- Zheng YY, Ma YT, Zhang JY, Xie X. COVID-19 and the cardiovascular system. Nat Rev Cardiol. 2020 May;17(5):259-260. doi: 10.1038/s41569-020-0360-5. PMID: 32139904; PMCID: PMC7095524.
- Xu L, Liu J, Lu M, Yang D, Zheng X. Liver injury during highly pathogenic human coronavirus infections. Liver Int. 2020 May;40(5):998-1004. doi: 10.1111/liv.14435. Epub 2020 Mar 30. PMID: 32170806; PMCID: PMC7228361.
- Yang Y, Guo F, Zhao W, Gu Q, Huang M, Cao Q, Shi Y, Li J, Chen J, Yan J, Jin Z, Wang X, Deng Y, Sun L, Cai H, Huang J, Zheng Y, Li W, Liu A, Chen B, Zhou M, Qiu H, Slutsky AS. Novel avian-origin influenza A (H7N9) in critically ill patients in China*. Crit Care Med. 2015 Feb;43(2):339-45. doi: 10.1097/CCM.0000000000000695. PMID: 25365721.
- Zhou L, Liu HG. [Early detection and disease assessment of patients with novel coronavirus pneumonia]. Zhonghua Jie He He Hu Xi Za Zhi. 2020 Feb 5;43(0):E003. Chinese. doi: 10.3760/cma.j.issn.1001-0939.2020.0003. Epub ahead of print. PMID: 32023686.
- Paules C, Subbarao K. Influenza. Revue De Linfirmière 2017, 390:39-44.
- Meng GG, Luo WF, Li ZL, Pan SF, Lu PX. Clinical and imaging characteristics of novel influenza A (H1N1) infection in 31 severe and critically ill patients. Radiologic Practice 2010; 25(29): 961-964.
- Keilman LJ. Seasonal Influenza (Flu). Nurs Clin North Am. 2019 Jun;54(2):227-243. doi: 10.1016/j.cnur.2019.02.009. PMID: 31027663.
- Schulert GS, Zhang M, Fall N, Husami A, Kissell D, Hanosh A, Zhang K, Davis K, Jentzen JM, Napolitano L, Siddiqui J, Smith LB, Harms PW, Grom AA, Cron RQ. Whole-Exome Sequencing Reveals Mutations in Genes Linked to Hemophagocytic Lymphohistiocytosis and Macrophage Activation Syndrome in Fatal Cases of H1N1 Influenza. J Infect Dis. 2016 Apr 1;213(7):1180-8. doi: 10.1093/infdis/jiv550. Epub 2015 Nov 23. PMID: 26597256; PMCID: PMC4779301.
- Tran TH, Nguyen TL, Nguyen TD, Luong TS, Pham PM, Nguyen vV, Pham TS, Vo CD, Le TQ, Ngo TT, Dao BK, Le PP, Nguyen TT, Hoang TL, Cao VT, Le TG, Nguyen DT, Le HN, Nguyen KT, Le HS, Le VT, Christiane D, Tran TT, Menno de J, Schultsz C, Cheng P, Lim W, Horby P, Farrar J; World Health Organization International Avian Influenza Investigative Team. Avian influenza A (H5N1) in 10 patients in Vietnam. N Engl J Med. 2004 Mar 18;350(12):1179-88. doi: 10.1056/NEJMoa040419. Epub 2004 Feb 25. PMID: 14985470.
- Harrison AG, Lin T, Wang P. Mechanisms of SARS-CoV-2 Transmission and Pathogenesis. Trends Immunol. 2020 Dec;41(12):1100-1115. doi: 10.1016/j.it.2020.10.004. Epub 2020 Oct 14. PMID: 33132005; PMCID: PMC7556779.
- Rello J, Storti E, Belliato M, Serrano R. Clinical phenotypes of SARS-CoV-2: implications for clinicians and researchers. Eur Respir J. 2020 May 21;55(5):2001028. doi: 10.1183/13993003.01028-2020. PMID: 32341111; PMCID: PMC7236837.
- Gattinoni L, Chiumello D, Caironi P, Busana M, Romitti F, Brazzi L, Camporota L. COVID-19 pneumonia: different respiratory treatments for different phenotypes? Intensive Care Med. 2020 Jun;46(6):1099-1102. doi: 10.1007/s00134-020-06033-2. Epub 2020 Apr 14. PMID: 32291463; PMCID: PMC7154064.
- Jain A, Doyle DJ. Stages or phenotypes? A critical look at COVID-19 pathophysiology. Intensive Care Med. 2020 Jul;46(7):1494-1495. doi: 10.1007/s00134-020-06083-6. Epub 2020 May 18. PMID: 32424481; PMCID: PMC7232601.
- Iba T, Levy JH, Levi M, Connors JM, Thachil J. Coagulopathy of Coronavirus Disease 2019. Crit Care Med. 2020 Sep;48(9):1358-1364. doi: 10.1097/CCM.0000000000004458. PMID: 32467443; PMCID: PMC7255402.
- Tang N, Li D, Wang X, Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost. 2020 Apr;18(4):844-847. doi: 10.1111/jth.14768. Epub 2020 Mar 13. PMID: 32073213; PMCID: PMC7166509.
- Cui S, Chen S, Li X, Liu S, Wang F. Prevalence of venous thromboembolism in patients with severe novel coronavirus pneumonia. J Thromb Haemost. 2020 Jun;18(6):1421-1424. doi: 10.1111/jth.14830. Epub 2020 May 6. PMID: 32271988; PMCID: PMC7262324.
- McGonagle D, Sharif K, O'Regan A, Bridgewood C. The Role of Cytokines including Interleukin-6 in COVID-19 induced Pneumonia and Macrophage Activation Syndrome-Like Disease. Autoimmun Rev. 2020 Jun;19(6):102537. doi: 10.1016/j.autrev.2020.102537. Epub 2020 Apr 3. PMID: 32251717; PMCID: PMC7195002.
- Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020 May;46(5):846-848. doi: 10.1007/s00134-020-05991-x. Epub 2020 Mar 3. Erratum in: Intensive Care Med. 2020 Apr 6;: PMID: 32125452; PMCID: PMC7080116.
- AminJafari A, Ghasemi S. The possible of immunotherapy for COVID-19: A systematic review. Int Immunopharmacol. 2020 Jun;83:106455. doi: 10.1016/j.intimp.2020.106455. Epub 2020 Apr 2. PMID: 32272396; PMCID: PMC7128194.
- Shanmugaraj B, Siriwattananon K, Wangkanont K, Phoolcharoen W. Perspectives on monoclonal antibody therapy as potential therapeutic intervention for Coronavirus disease-19 (COVID-19). Asian Pac J Allergy Immunol. 2020 Mar;38(1):10-18. doi: 10.12932/AP-200220-0773. PMID: 32134278.
- Zhou G, Zhao Q. Perspectives on therapeutic neutralizing antibodies against the Novel Coronavirus SARS-CoV-2. Int J Biol Sci 2020, 16:1718-1723.
- Zhou B, Zhong N, Guan Y. Treatment with convalescent plasma for influenza A (H5N1) infection. N Engl J Med. 2007 Oct 4;357(14):1450-1. doi: 10.1056/NEJMc070359. PMID: 17914053.
- Hung IF, To KK, Lee CK, Lee KL, Chan K, Yan WW, Liu R, Watt CL, Chan WM, Lai KY, Koo CK, Buckley T, Chow FL, Wong KK, Chan HS, Ching CK, Tang BS, Lau CC, Li IW, Liu SH, Chan KH, Lin CK, Yuen KY. Convalescent plasma treatment reduced mortality in patients with severe pandemic influenza A (H1N1) 2009 virus infection. Clin Infect Dis. 2011 Feb 15;52(4):447-56. doi: 10.1093/cid/ciq106. Epub 2011 Jan 19. PMID: 21248066; PMCID: PMC7531589.
- Hui DS, Lee N, Chan PK, Beigel JH. The role of adjuvant immunomodulatory agents for treatment of severe influenza. Antiviral Res. 2018 Feb;150:202-216. doi: 10.1016/j.antiviral.2018.01.002. Epub 2018 Jan 8. PMID: 29325970; PMCID: PMC5801167.
- Cortegiani A, Ingoglia G, Ippolito M, Giarratano A, Einav S. A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19. J Crit Care. 2020 Jun;57:279-283. doi: 10.1016/j.jcrc.2020.03.005. Epub 2020 Mar 10. PMID: 32173110; PMCID: PMC7270792.
- Gao J, Tian Z, Yang X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends. 2020 Mar 16;14(1):72-73. doi: 10.5582/bst.2020.01047. Epub 2020 Feb 19. PMID: 32074550.
- Wang M, Cao R, Zhang L, Yang X, Liu J et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 2020; 30:269-271.
- Luo H, Tang QL, Shang YX, Liang SB, Yang M, Robinson N, Liu JP. Can Chinese Medicine Be Used for Prevention of Corona Virus Disease 2019 (COVID-19)? A Review of Historical Classics, Research Evidence and Current Prevention Programs. Chin J Integr Med. 2020 Apr;26(4):243-250. doi: 10.1007/s11655-020-3192-6. Epub 2020 Feb 17. PMID: 32065348; PMCID: PMC7088641.
- Zhu FC, Li YH, Guan XH, Hou LH, Wang WJ et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet 2020; 395:1845-1854.
- National health commission of the people's republic of China.
- Wang D, Yin Y, Hu C, Liu X, Zhang X, Zhou S, Jian M, Xu H, Prowle J, Hu B, Li Y, Peng Z. Clinical course and outcome of 107 patients infected with the novel coronavirus, SARS-CoV-2, discharged from two hospitals in Wuhan, China. Crit Care. 2020 Apr 30;24(1):188. doi: 10.1186/s13054-020-02895-6. PMID: 32354360; PMCID: PMC7192564.
- Grasselli G, Zangrillo A, Zanella A, Antonelli M, Cabrini L, Castelli A, Cereda D, Coluccello A, Foti G, Fumagalli R, Iotti G, Latronico N, Lorini L, Merler S, Natalini G, Piatti A, Ranieri MV, Scandroglio AM, Storti E, Cecconi M, Pesenti A; COVID-19 Lombardy ICU Network. Baseline Characteristics and Outcomes of 1591 Patients Infected With SARS-CoV-2 Admitted to ICUs of the Lombardy Region, Italy. JAMA. 2020 Apr 28;323(16):1574-1581. doi: 10.1001/jama.2020.5394. Erratum in: JAMA. 2021 May 25;325(20):2120. PMID: 32250385; PMCID: PMC7136855.
- Zheng Z, Yao Z, Wu K, Zheng J. Patient follow-up after discharge after COVID-19 pneumonia: Considerations for infectious control. J Med Virol. 2020 Nov;92(11):2412-2419. doi: 10.1002/jmv.25994. Epub 2020 Aug 21. PMID: 32383776; PMCID: PMC7267672.
- Su JW, Wu WR, Lang GJ, Zhao H, Sheng JF. Transmission risk of patients with COVID-19 meeting discharge criteria should be interpreted with caution. J Zhejiang Univ Sci B 2020; 21:408-410.
- Li Y, Hu Y, Zhang X, Yu Y, Li B, Wu J, Wu Y, Xia X, Xu J. [Follow-up testing of viral nucleic acid in discharged patients with moderate type of 2019 coronavirus disease (COVID-19)]. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2020 May 25;49(2):270-274. Chinese. doi: 10.3785/j.issn.1008-9292.2020.03.11. PMID: 32222122; PMCID: PMC8800664.
- He Z. What further should be done to control COVID-19 outbreaks in addition to cases isolation and contact tracing measures? BMC Med. 2020 Mar 13;18(1):80. doi: 10.1186/s12916-020-01551-8. PMID: 32164708; PMCID: PMC7069035.
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.
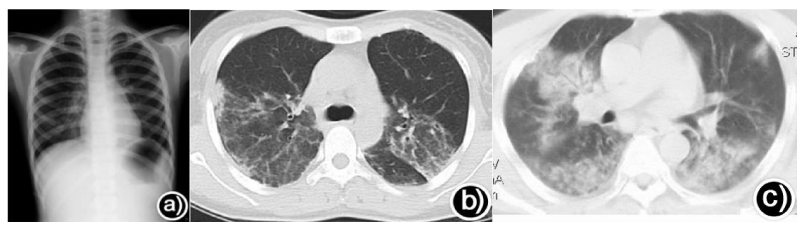
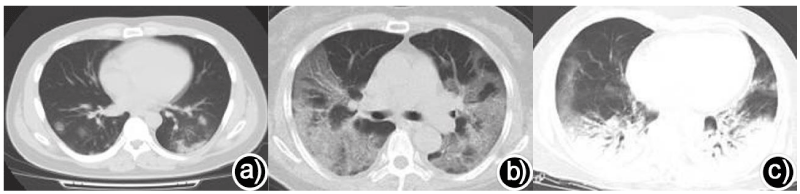

 Save to Mendeley
Save to Mendeley
