International Journal of Pharmaceutical Sciences and Developmental Research
In silico phytochemicals analysis as inhibitors of the SARS-COV-2 main protease
Ekaterina Serikova1,2£, Victor Gustavo Oliveira Evangelho1£, Marianna Kremenevskaya2, Camila Ferreira Mattos3, Juliana Silva Novais3, Marcos Vinicius Santana1, Carlos Rangel Rodrigues1,3, Reinaldo Barros Geraldo1* and Helena Carla Castro3*
2Faculty of Biotechnologies (BioTech), ITMO University, Lomonosov st. 9, 191002 St. Petersburg, Russia
3Postgraduate Pathology Program, Fluminense Federal University, Antônio Pedro University Hospital, 24033-900, Niterói-RJ, Brazil
£These co-authors share the first authorship
Reinaldo Barros Geraldo, PhD, Postgraduate in Science and Biotechnology, Fluminense Federal University, 24033-900, Niterói, Rio de Janeiro, Brazil, E-mail: reinaldobgeraldo@gmail.com
Cite this as
Serikova E, Oliveira Evangelho VG, Kremenevskaya M, Geraldo RB, Castro HC, et al. (2022) In silico phytochemicals analysis as inhibitors of the SARS-COV-2 main protease. Int J Pharm Sci Dev Res 8(1): 038-045. DOI: 10.17352/ijpsdr.000041Copyright License
© 2022 Serikova E, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Background: The world population's full immunization with vaccines against SARS-CoV-2 is still challenging. Therefore, more research must be needed to find an active antiviral drug against the virus, including new mutated strains.
Results: Therefore, this research analyzes 35 natural compounds isolated from various plants against SARS-CoV-2 main protease (Mpro) using an in silico strategy. According to the results, it was possible to identify promising molecules using a molecular docking strategy. Furthermore, the results showed that the interaction of these molecules with protease-specific residues, including (2S)-Eriodictyol 7-O-(6''-O-galloyl)-beta-D-glucopyranoside (Trp207, Ser284, and Glu288), Hypericin (Glu166, Arg188, and Thr190), Calceolarioside B (Gly143, Ser144, Cys145, Glu166, Arg188, and Gln192), Epicatechin (Ser144, His163, and Leu167) and Myricitrin (Thr190) with ΔG was -8.5, -9.6, -8.5, -9.3 and -9.3 kcal/mol, respectively. In addition, analyzing all compounds for their ADME properties shows that compounds present an excellent pharmacokinetic profile.
Conclusion: In conclusion, the results of this study indicated that these major natural compounds can be considered potential inhibitors of Mpro and should be further explored in vitro and in vivo in accordance with our data.
Abbreviations
SARS-CoV-2: Severe Acute Respiratory Syndrome Coronavirus 2; Mpro: Main protease; ADME: Absorption, Distribution, Metabolism, and Excretion; Trp: Tryptophane; Ser: Serine; Glu: Glutamic acid; Arg: Arginine; Thr: Threonine; Gly: Glycine; Cys: Cysteine; Gln: Glutamine; Leu: Leucine; ΔG: Gibbs free energy; kcal: kilo calories; mol: mole; VOC: Variants of Concern; MMFF94: Merck Molecular Force Field 94; 3D: Three Dimensional; PDB ID: Protein Data Bank Identification; mol2: portable representation of an SYBYL molecule; SMILE: Simplified Molecular-Input Line-Entry System; FDA: U.S. Food and Drug Administration; ADMET: Absorption, Distribution, Metabolism, Excretion and Toxicity; Å2: Variation of the mean accessible surface area; H-bond: Hydrogen bond; logP: logarithm (base 10) of the partition coefficient P; Log Po/w: Logarithm (base 10) of the octanol/water partition coefficient
Background
Although many countries have reviewed restrictive measures and have relaxed the use of masks [1], some countries do not have a fully immunized population. This scenario could highlight the middle- or low-income countries with few resources to acquire the developed immunizers and lack donations from richer countries [2,3]. In addition, in some countries, the population is waiting for newer vaccine applications as a booster against COVID-19 [4-6]. When the pandemic started, pharmaceutical enterprises suggested several drugs as treatments. However, they presented significantly less effectiveness than previous data suggested in clinical trials [7]. In addition, coronavirus can rapidly mutate, which may demand new effective vaccines shortly [8].
In this context, the identified variants have elevated the race for new drugs and vaccines against important VOC, such as alpha (B.1.1.7), beta (B. 1.351), gamma (P.1), and delta (B. 1.617.2). Moreover, the low vaccine adherence in several countries promoted the development of new variants. This scenario is the case of the omicron Variant (B.1.1.529), whose dissemination caused outbreaks in several countries in early 2022. Moreover, again, ringing the warnings, the fine line among new variants, and the risk of resistance to already developed vaccines [9,10]. In particular, the omicron variant has many mutations in the Spike protein. These mutations may contribute to the low efficacy of current vaccines and provide virus evasion and already immunized people [11].
According to recent studies, COVID-19 vaccines approved to date produce serum IgG1 but leave nasal epithelium and upper respiratory tract unprotected. As a result, they allow the SARS-CoV-2 infection (i.e., not providing mucosal immunity) [12]. What makes necessary new pathways for COVID-19 infection therapies? In this instance, natural or synthetic compounds are an awesome font to discover new antiviral drugs. Plant compounds include an extensive list of biologically active molecules. Several of them have antiviral activity [13]. Thus, they are a good starting point for the search against those interactions with virus proteins.
The SARS-CoV-2 main protease (Mpro) is a promising drug target. This protein plays a crucial role in viral replication and transcription [14]. Mpro is conserved among coronaviruses, sharing ~76% sequence similarity with SARS-CoV-1 Mpro. In addition, there is no human host cell protease with similar substrate specificity [15].
At last, molecular docking is an excellent methodology for predicting their protein-ligand interaction. Several endpoints, such as interactive activity, interaction energy, and ligand-specific interaction region, are essential in that approach. Consequently, molecular docking studies could help model the ligand-receptor interaction to identify potential SARS-CoV-2 Mpro pharmacophore sites. Herein, it aims to test 35 natural compounds as potential inhibitors of the SARS-CoV-2 major protease. In addition, it analyzed the computational pharmacokinetic parameters and predicted their toxicological effects on humans. Lastly, this study resembles information on a drug structure through standard and drug similarity.
Methods
Molecular docking studies
Molecular docking was performed via the Chimera tool, using the AutodockVina tool to predict the protein-ligand interaction described elsewhere [16]. Docking process possessing parameter of maximum reiteration of 1500, maximum population size 50, Grid solution 0.2 having a binding affinity, the protein and ligands were evaluated on the following confirmation of the Internal Electrostatic interaction (Internal ES), sp2-sp2 torsions and internal hydrogen bond interaction. The binding site is defined as the first cavity possessing high volume. A post dock study involves energy minimization and H-bond optimization. Setting of Simplex Evolution at max steps 300 and neighbor distance faster 1.00 [16]. The method for docking calculation consists of using the MMFF94 parameter as a Geometry Optimization Method, where a force field aims to minimize the energy of the ligand. The solvation parameters are set by AutoDockVina concurrently with the docking simulations [17]. The docking validation was following the instructions presented elsewhere [18].
Ligands preparation
In this work, there are evaluated 35 phytochemical components from various plants using a molecular docking protocol. Some compounds have antiviral activities described yet: Myricitrin, Methyl rosemary, 5,7,3',4'-tetrahydroxy-2'-(3,3-dimethylallyl) isoflavone, (2S)-Eriodictyol 7-O-(6"-O-galloyl)-beta-D-glycopyranoside, calceolarioside B, Myricetin 3-O-beta-D-glucopyranoside, Licoleafol, Amarantine, Berberine, Caffeine, Capsaicin, Embelin, Emodin, Gossypol, Hypericin, Luteolin, Oxyacanthine, Sanguinarian, Crocin, Digitoxigenin, β-eudesmol, Kaempferol, Quercetin, Naringenin, Oleuropein, Catechin, Curcumin, Epicatechin-gallate, Zingerol, Gingerol, Allicin, Resveratrol, Pterostilbene, Pinosilvin, Piceatannol. The ligand structures were taken from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) in mol2 format for docking analyses.
Protease preparation
Mpro three-dimensional (3D) structure was chosen for the in silico test against those natural compounds. This structure is available at the Protein Databank in a complex with an inhibitor (PDB ID: 6LU7). The 3D structure of the SARS-CoV-2 main protease was downloaded from Protein Data Bank (https://www.rcsb.org). The 6LU7 protein is a homodimer and contains 306 amino acid residues, and two chains, named A and B. Chain A was used for macromolecule preparation. As performed in Sahin, et al., the water molecules were removed, and nonpolar hydrogen molecules were added to the protein structure in the Python molecule viewer setting of mgltools, and a [17;19]. The complexed inhibitor was removed using UCSF Chimera 1.15, and the apoprotein was saved in PDB format [20]. The protein was minimized using SwissProt PDBviewer 4.1 as described elsewhere [21].
ADME analysis
Using the canonical SMILES chemical nomenclature of each ligand under consideration, it performed Absorption, Distribution, Metabolism, and Excretion (ADME) prediction. Then, the molecules are uploaded in SMILES format to the SwissADME server (http://www.swissadme.ch). We subsequently surveyed HIV protease inhibitors, already authorized by the FDA as positive controls. Thus, with this approach, some parameters were established among the tested molecules and some inhibitors already on the market.
Results
Molecular docking studies
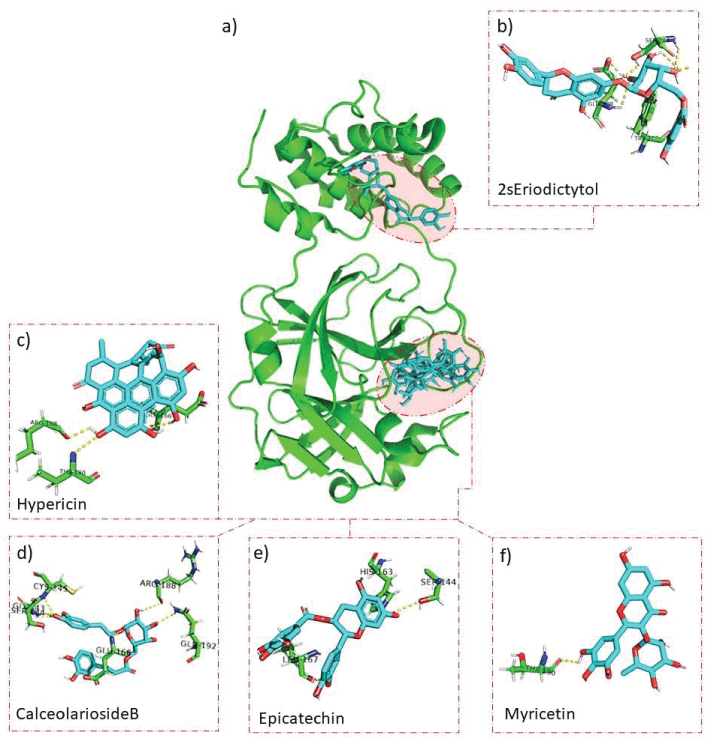
Molecular docking was performed for 35 phytochemical components against Mpro using UCSF Chimera 1.15 to identify molecular interactions between the protein active site and ligands. The binding energy (ΔG) in these complexes' best poses is shown and compared in Figure 1. The best ligands were those with the highest binding energy, including Myricitrin, Calceolarioside B, Hypericin, Epicatechin-gallate, (2S)-Eriodictyol 7-O-(6''-O-galloyl)-beta-D-glucopyranoside and Calceolarioside B.
The interaction analysis of these natural molecules with the virus protease revealed specific residues, including when interacting with the (2S)-Eriodictyol 7-O-(6''-O-galloyl)-beta-D-glucopyranoside (Trp207, Ser284, and Glu288), Hypericin (Glu166, Arg188, and Thr190), Calceolarioside B (Gly143, Ser144, Cys145, Glu166, Arg188, and Gln192), Epicatechin (Ser144, His163, and Leu167) and Myricitrin (Thr190) (Figure 2). These data may indicate that these compounds may be a feasible option to explore against some specific mutants of this coronavirus.
ADMET prediction
The analysis of the physical and chemical properties of all molecules, their lipophilicity, and drug likeliness are shown in Table 1. The rule of five (RO5) predicts that good absorption is more likely when the molecular weight is lower than 500, the value of the topological polar surface area is in the range of 20-140 Å2, and the value of Molecular Refractivity in the range 40-130 and there are less than 5 H-bond donors and 10 H-bond acceptors.
Discussion
Molecular docking is a fundamental tool for intermolecular interaction prediction. These results might direct subsequent studies. Herein, the lowest docking score determined the preferred best ligand as it indicates more excellent stability of the binding [22]. Based on molecular docking results among the phytochemicals with the Mpro active site, six compounds exhibited better binding efficacy against the target. Compared to the other phytochemicals, these compounds are Myricitrin, (2S)-Eriodictyol 7-O-(6''-O-galloyl)-beta-D-glucopyranoside, Calceolarioside B, Myricetin 3-O-beta-D-glucopyranoside, Hypericin, and Epicatechin-gallate.
In another inhibitor analysis, the peptide inhibitor N3 and ∝-ketamine interacted via hydrogen bonds with residues in the protease binding site. The interactions happen among the compounds and Thr190, His163, Cys145, Glu166, and Gly143 protein residues. These residues are also the interaction targets of the ligands tested in our study Hypericin, Epicatechin, Myricitrin, and Calceolarioside B [23].
Another study shares these interaction residues. Interactions with Remdesivir (Cys145, Glu166, Gln192, and Thr190), GS441524 (Glu166, Gln192, and Thr190), Arbidol (His163, Glu166, and Leu167), Glycyrrhizin (Glu166, Cys145, and Ser144) and native co-crystal ligand (O6K) (Glu166 and His163) were found to interact with these regions of the protease [24] specifically. Mpro has three domains, consisting of residues 8-101 (Domain I), 102-184 (Domain II), and 201-303 (Domain III) [25]. This result indicates that the most relevant ligands in our research are between domains I and II of Mpro. Our data indicate a ΔG close to or higher than those found in other works that tested other natural producers, such as the generous Aloe [26], Xylopic acid, Resveratrol, Ellagic acid, Kaempferol, and Quercetin [27] and diterpenoids and bioflavonoids of TorreyaNucifera leaves [28].
This rule describes molecular properties that are important for the pharmacokinetics of a drug in the human body. This rule can be used when creating drugs. The active drug is gradually optimized to increase the activity and selectivity of the compound and to ensure that the physicochemical properties inherent in compounds obeying the Lipinski rule are maintained.
Bioavailability Score is based on a probability value of a molecule to have an optimum profile of permeability and bioavailability, where 0.55 indicates the obedience of the Lipinski rule of five. A molecule can be considered a good drug if all the computational filters of SwissADME - Lipinski, Ghose, Veber, Egan, and Mueggehave no violations.
Myricitrin, Calceolarioside B, and Myricetin 3-O-beta-D-glucopyranoside have several H-bond acceptors equal to 12, 11 and 13, respectively, which is more than ten acceptable. The molecular weight of (2S)-Eriodictyol 7-O-(6''-O-galloyl)-beta-D-glucopyranoside is not lower than 500 g/mol and equal to 602.50 g/mol as well as Hypericin. Epicatechin-gallate showed the value of topological polar surface area equal to 177.14 Å2, which is not in the excellent range of 20-140 Å2.
The lipophilicity study revealed that values of Log Po/w (Consensus Log Po/w) of Gossypol and Oxyacanthine are out of the acceptable five and equal to 5.04 and 5.12, respectively. In addition, Myricitrin, Myricetin 3-O-beta-D-glucopyranoside, Amaranthine, and Crocin have negative lipophilicity values, which is unfavorable.
Hypericin is an anthraquinone derivative naturally found in the yellow flower of Hypericumperforatum with an antidepressant, potential antiviral, antineoplastic and immunostimulating activities [29]. It has the highest docking score (-9.6 Kcal/mol) with the main coronavirus protease among all the phytochemicals.
The use of molecules available in nature offers an advantage for the bioavailability of these compounds. Considering the results obtained from molecular docking studies, the phytochemicals Myricitrin, (2S)-Eriodictyol 7-O-(6''-O-galloyl)-beta-D-glucopyranoside, Calceolarioside B, Myricetin 3-O-beta-D-glucopyranoside, Hypericin, and Epicatechin-gallate can be considered for exploring in COVID-19 treatment. These phytochemicals have shown comparable coronavirus main protease in silico inhibitory.
Flavonoid Myricitrin is found in vegetables, fruits, nuts, berries, herbs, plants, tea, wine, and medicinal plants. Myricitrin is nitric oxide (NO) and protein kinase C (PKC) inhibitor that has central nervous system activity, including anxiolytic-like action [30] docked with a target with a docking score of -9.3 Kcal/mol.
Myricetin 3-O-beta-D-glucopyranoside is a compound with chemotherapeutic, chemopreventive, and antiangiogenic properties [31], a naturally occurring plant metabolite found in several food items such as blackcurrant, typical grape, highbush blueberry, and tea docked with the target with a docking score of -9.3 Kcal/mol.
Epicatechin-gallate, a flavonoid present in brewed black, green, and oolong teas, cherries, and strawberries inhibit the growth of cancer cells, and gas anti-inflammatory effects show a docking score of -9.3 Kcal/mol [32].
(2S)-Eriodictyol 7-O-(6''-O-galloyl)-beta-D-glucopyranoside isolated from the leaves of Acer mono possessing antioxidant activity [33], has binding efficacy with a target with a docking score of -8.5 Kcal/mol. Calceolarioside B, a natural product found in Lepisoruscontortus [34] binds with a docking score of -8.5 Kcal/mol.
Compounds conforming to Lipinski's rule are less intensively consumed during clinical trials and therefore have an increased likelihood of entering the market. In addition, according to Lipinski's rule, the molecular weight should be less than 500 g/mol. This characteristic is necessary to transport the ligand into cells easily. The octanol-water distribution coefficient (logP) characterizes the ability to transport drugs through cell membranes, determining their absorption and distribution in various body systems. A negative logP value means that the compound has a higher affinity to the aqueous phase; when logP = 0, the compound is equally distributed between the lipid and aqueous phases; a positive logP value means a higher concentration of the compound in the lipid phase [35-38].
Lipinsky's Rule of Five perspectives can conclude that none of the phytochemicals above has properties that make it an orally active drug. Thus, they may require different formulations, including nanotechnology or strategy/administration routes (e.g., use of injection). However, compared with the FDA-approved protease inhibitor positive controls, they may still have a good profile that should be evaluated by in vitro and in vivo assays.
Based on these results, we can highlight that in a globalized world, where territorial barriers are easily broken by airplane and boat trips that daily take hundreds of individuals from countries with low vaccination coverage to other locations of the globe, igniting an alert for the importance of investments in pharmacological alternatives against SARS-Cov-2.
Conclusion
With molecular docking, the screening results in 35 natural products as possible competitive inhibitors of SARS-Cov-2 main protease. In addition, molecular docking studies disclosed the high binding affinities of (2S)-Eriodictyol 7-O-(6''-O-galloyl)-beta-D-glucopyranoside, Hipericina, Calceolarioside B, Epicatechin and Myricitrin.
Physicochemical parameters also showed promising properties of drug-likeness. The current study results conclude that the complexity of natural compounds can be considered a preventive measure to reduce or prevent the effects of COVID-19. Further research should be conducted to verify the potency and safety of these compounds in the treatment of COVID-19.
Declarations
Contributions: ES, VGOE, RBG, and HCC performed the research and manuscript writing. JSN assisted in the bibliographic organization and carried out the writing of the manuscript. ES, MVS, VGOE performed the computational experimental work, analyzed the data, created the figures and tables and wrote the manuscript. RBG, HCC, MK, and CRR were responsible for the final review of the research and approval of the final content of the manuscript. All authors read and approved the final manuscript
Funding: This work was funded by CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), with master and doctorate scholarships in the postgraduate Program in Science and Biotechnology, Universidade Federal Fluminense.
The authors would like to thank CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), FAPERJ (Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro), UFF (Universidade Federal Fluminense) and UFRJ (Universidade Federal do Rio de Janeiro), Brazil.
- Pan American Health Organization. Too soon to lower our guard, warns PAHO Director as the pandemic reaches the two-year mark. Accessed April 1, 2022. https://www.paho.org/en/news/9-3-2022-too-soon-lower-our-guard-warns-paho-director-pandemic-reaches-two-year-mark
- Tagoe ET, Sheikh N, Morton A, Nonvignon J, Sarker AR, Williams L, Megiddo I. COVID-19 Vaccination in Lower-Middle Income Countries: National Stakeholder Views on Challenges, Barriers, and Potential Solutions. Front Public Health. 2021 Aug 6;9:709127. doi: 10.3389/fpubh.2021.709127. PMID: 34422750; PMCID: PMC8377669.
- Ekström AM, Tomson G, Wanyenze RK, Bhutta ZA, Kyobutungi C, Binagwaho A, Ottersen OP. Addressing production gaps for vaccines in African countries. Bull World Health Organ. 2021 Dec 1;99(12):910-912. doi: 10.2471/BLT.21.287381. Epub 2021 Nov 17. PMID: 34866689; PMCID: PMC8640685.
- Shekhar R, Garg I, Pal S, Kottewar S, Sheikh AB. COVID-19 Vaccine Booster: To Boost or Not to Boost. Infect Dis Rep. 2021 Oct 28;13(4):924-929. doi: 10.3390/idr13040084. PMID: 34842753; PMCID: PMC8628913.
- García-Botella A, García-Lledó A, Gómez-Pavón J, González Del Castillo J, Hernández-Sampelayo T, Martín-Delgado MC, Martín Sánchez FJ, Martínez-Sellés M, Molero García JM, Moreno Guillén S, Rodríguez-Artalejo FJ, Ruiz-Galiana J, Cantón R, De Lucas Ramos P, Bouza E. Booster or additional vaccination doses in patients vaccinated against COVID-19. Rev Esp Quimioter. 2022 Apr;35(2):105-114. doi: 10.37201/req/149.2021. Epub 2021 Nov 15. PMID: 34775740; PMCID: PMC8972704.
- The Lancet Infectious Diseases. COVID-19 vaccine equity and booster doses. Lancet Infect Dis. 2021 Sep;21(9):1193. doi: 10.1016/S1473-3099(21)00486-2. Epub 2021 Aug 13. PMID: 34391506; PMCID: PMC8360703.
- BBC. Sinovac: Brazil results show the Chinese vaccine is 50.4% effective. Bbc. Published online 2021.
- Jiang Y, Yin W, Xu HE. RNA-dependent RNA polymerase: Structure, mechanism, and drug discovery for COVID-19. Biochem Biophys Res Commun. 2021 Jan 29;538:47-53. doi: 10.1016/j.bbrc.2020.08.116. Epub 2020 Sep 4. PMID: 32943188; PMCID: PMC7473028.
- World Health Organization. Accessed November 29, 2021,https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern
- Toovey OTR, Harvey KN, Bird PW, Tang JWW. Introduction of Brazilian SARS-CoV-2 484K.V2 related variants into the UK. J Infect. 2021 May;82(5):e23-e24. doi: 10.1016/j.jinf.2021.01.025. Epub 2021 Feb 3. PMID: 33548358; PMCID: PMC7857057.
- Callaway E. Heavily mutated Omicron variant puts scientists on alert. Nature. 2021 Dec;600(7887):21. doi: 10.1038/d41586-021-03552-w. PMID: 34824381.
- Ratcliffe NA, Castro HC, Paixão IC, Evangelho VGO, Azambuja P, Mello CB. Nasal therapy-The missing link in optimising strategies to improve prevention and treatment of COVID-19. PLoS Pathog. 2021 Nov 24;17(11):e1010079. doi: 10.1371/journal.ppat.1010079. PMID: 34818380; PMCID: PMC8612505.
- Tseliou M, Pirintsos SA, Lionis C, Castanas E, Sourvinos G. Antiviral effect of an essential oil combination derived from three aromatic plants (Coridothymuscapitatus (L.) Rchb. f., Origanumdictamnus L. and Salvia fruticosa Mill.) against viruses causing infections of the upper respiratory tract. Journal of Herbal Medicine. 2019;17-18 March 2018;100288. doi:10.1016/j.hermed.2019;100288
- Dai J, Han R, Xu Y, Li N, Wang J, Dan W. Recent progress of antibacterial natural products: Future antibiotics candidates. Bioorg Chem. 2020 Aug;101:103922. doi: 10.1016/j.bioorg.2020.103922. Epub 2020 May 20. PMID: 32559577.
- Ullrich S, Nitsche C. The SARS-CoV-2 main protease as drug target. Bioorg Med Chem Lett. 2020 Sep 1;30(17):127377. doi: 10.1016/j.bmcl.2020.127377. Epub 2020 Jul 2. PMID: 32738988; PMCID: PMC7331567.
- Shukla P, Khandelwal R, Sharma D, Dhar A, Nayarisseri A, Singh SK. Virtual Screening of IL-6 Inhibitors for Idiopathic Arthritis. Bioinformation. 2019 Feb 28;15(2):121-130. doi: 10.6026/97320630015121. PMID: 31435158; PMCID: PMC6677908.
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009 Dec;30(16):2785-91. doi: 10.1002/jcc.21256. PMID: 19399780; PMCID: PMC2760638.
- CS, S DK, Ragunathan V, Tiwari P, A S, P BD. Molecular docking, validation, dynamics simulations, and pharmacokinetic prediction of natural compounds against the SARS-CoV-2 main-protease. J Biomol Struct Dyn. 2022 Feb;40(2):585-611. doi: 10.1080/07391102.2020.1815584. Epub 2020 Sep 8. PMID: 32897178; PMCID: PMC7573242.
- Sahin S, Calapoglu, Ozmen FI. Didemnins Inhibit COVID-19 Main Protease (Mpro). Biointerface Research in Applied Chemistry. 2021; 11(1):8204-8209. https://doi.org/10.33263/BRIAC111.82048209
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004 Oct;25(13):1605-12. doi: 10.1002/jcc.20084. PMID: 15264254.
- de Castro CC, Costa PS, Laktin GT, de Carvalho PH, Geraldo RB, de Moraes J, Pinto PL, Couri MR, Pinto Pde F, Da Silva Filho AA. Cardamonin, a schistosomicidal chalcone from Piper aduncum L. (Piperaceae) that inhibits Schistosoma mansoni ATP diphosphohydrolase. Phytomedicine. 2015 Sep 15;22(10):921-8. doi: 10.1016/j.phymed.2015.06.009. Epub 2015 Jul 4. PMID: 26321741.
- Krovat E, Steindl T, Langer T. Recent Advances in Docking and Scoring. Current Computer Aided-Drug Design. 2006;1(1):93-102. doi:10.2174/1573409052952314
- Choudhary MI, Shaikh M, Tul-Wahab A, Ur-Rahman A. In silico identification of potential inhibitors of key SARS-CoV-2 3CL hydrolase (Mpro) via molecular docking, MMGBSA predictive binding energy calculations, and molecular dynamics simulation. PLoS One. 2020 Jul 24;15(7):e0235030. doi: 10.1371/journal.pone.0235030. PMID: 32706783; PMCID: PMC7380638.
- Weng YL, Naik SR, Dingelstad N, Lugo MR, Kalyaanamoorthy S, Ganesan A. Molecular dynamics and in silico mutagenesis on the reversible inhibitor-bound SARS-CoV-2 main protease complexes reveal the role of lateral pocket in enhancing the ligand affinity. Sci Rep. 2021 Apr 1;11(1):7429. doi: 10.1038/s41598-021-86471-0. PMID: 33795718; PMCID: PMC8016996.
- Komatsu TS, Okimoto N, Koyama YM, Hirano Y, Morimoto G, Ohno Y, Taiji M. Drug binding dynamics of the dimeric SARS-CoV-2 main protease, determined by molecular dynamics simulation. Sci Rep. 2020 Oct 12;10(1):16986. doi: 10.1038/s41598-020-74099-5. PMID: 33046764; PMCID: PMC7550358.
- Abouelela ME, Assaf HK, Abdelhamid RA, Elkhyat ES, Sayed AM, Oszako T, Belbahri L, El Zowalaty AE, Abdelkader MSA. Identification of Potential SARS-CoV-2 Main Protease and Spike Protein Inhibitors from the Genus Aloe: An In Silico Study for Drug Development. Molecules. 2021 Mar 21;26(6):1767. doi: 10.3390/molecules26061767. PMID: 33801151; PMCID: PMC8004122.
- Oso BJ, Olaoye IF, Omeike SO. Molecular Docking and ADMET Prediction of Natural Compounds towards SARS Spike Glycoprotein-Human Angiotensin-Converting Enzyme 2 and SARS-CoV-2 Main Protease. Arch Razi Inst. 2021 Sep 1;76(3):453-459. doi: 10.22092/ari.2020.351202.1517. PMID: 34824739; PMCID: PMC8605841.
- Ghosh R, Chakraborty A, Biswas A, Chowdhuri S. Computer aided identification of potential SARS CoV-2 main protease inhibitors from diterpenoids and biflavonoids of Torreya nucifera leaves. J Biomol Struct Dyn. 2022 Apr;40(6):2647-2662. doi: 10.1080/07391102.2020.1841680. Epub 2020 Nov 3. PMID: 33140695; PMCID: PMC7663460.
- Murthy HN, Kim YS, Park SY, Paek KY. Hypericins: biotechnological production from cell and organ cultures. Appl Microbiol Biotechnol. 2014 Nov;98(22):9187-98. doi: 10.1007/s00253-014-6119-3. Epub 2014 Oct 10. PMID: 25301586.
- Jayasinghe L, Amarasinghe NR, Arundathie BG, Rupasinghe GK, Jayatilake NH, Fujimoto Y. Antioxidant flavonol glycosides from Elaeocarpus serratus and Filicium decipiens. Nat Prod Res. 2012;26(8):717-21. doi: 10.1080/14786419.2010.551514. Epub 2011 Sep 19. PMID: 21923561.
- Lin CY, Lin LC, Ho ST, Tung YT, Tseng YH, Wu JH. Antioxidant activities and phytochemicals of leaf extracts from 10 native rhododendron species in taiwan. Evid Based Complement Alternat Med. 2014;2014:283938. doi: 10.1155/2014/283938. Epub 2014 Jun 2. PMID: 24987425; PMCID: PMC4060324.
- Du GJ, Zhang Z, Wen XD, Yu C, Calway T, Yuan CS, Wang CZ. Epigallocatechin Gallate (EGCG) is the most effective cancer chemopreventive polyphenol in green tea. Nutrients. 2012 Nov 8;4(11):1679-91. doi: 10.3390/nu4111679. PMID: 23201840; PMCID: PMC3509513.
- Hu Q, Zhang DD, Wang L, Lou H, Ren D. Eriodictyol-7-O-glucoside, a novel Nrf2 activator, confers protection against cisplatin-induced toxicity. Food Chem Toxicol. 2012 Jun;50(6):1927-32. doi: 10.1016/j.fct.2012.03.059. Epub 2012 Mar 23. PMID: 22465804.
- Yang JH, Kondratyuk TP, Jermihov KC, Marler LE, Qiu X, Choi Y, Cao H, Yu R, Sturdy M, Huang R, Liu Y, Wang LQ, Mesecar AD, van Breemen RB, Pezzuto JM, Fong HH, Chen YG, Zhang HJ. Bioactive compounds from the fern Lepisorus contortus. J Nat Prod. 2011 Feb 25;74(2):129-36. doi: 10.1021/np100373f. Epub 2011 Jan 24. PMID: 21261296; PMCID: PMC3069126.
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001 Mar 1;46(1-3):3-26. doi: 10.1016/s0169-409x(00)00129-0. PMID: 11259830.
- Sen D, Debnath P, Debnath B, Bhaumik S, Debnath S. Identification of potential inhibitors of SARS-CoV-2 main protease and spike receptor from 10 important spices through structure-based virtual screening and molecular dynamic study. J Biomol Struct Dyn. 2022 Feb;40(2):941-962. doi: 10.1080/07391102.2020.1819883. Epub 2020 Sep 18. PMID: 32948116; PMCID: PMC7544938.
- Agrawal A, Jain N, Kumar N, Kulkarnu G. Molecural docking study to identify potential inhibitor of Covid-19 main protease enzyme: an in-silico approach. ChemRxiv Biological and Medicinal Chemistry 2020; DOI:10.26434/chemrxiv.121709
- Tahir Ul, Qamar M, Alqahtani SM, Alamri MA, Chen LL. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J Pharm Anal. 2020 Aug;10(4):313-319. doi: 10.1016/j.jpha.2020.03.009. Epub 2020 Mar 26. PMID: 32296570; PMCID: PMC7156227.
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.



 Save to Mendeley
Save to Mendeley
