International Journal of Pharmaceutical Sciences and Developmental Research
Analysis of the mechanism underlying the effects of cyclophosphamide against triple-negative breast cancer by an integrative bioinformatics approach
Maojing Wang1, Wenwen Li2, Ding Li3 and Zhiwu Han4*
2Department of Hematology, Qingdao women and children`s Hospital Qingdao, China
3Department of TCM, The Affiliated Hospital of Qingdao University, Qingdao, China
4Department of Pharmacy, The Affiliated Hospital of Qingdao University, Qingdao, China
Cite this as
Wang M, Li W, Li D, Han Z (2021) Analysis of the mechanism underlying the effects of cyclophosphamide against triple-negative breast cancer by an integrative bioinformatics approach. Int J Pharm Sci Dev Res 7(1): 006-015. DOI: 10.17352/ijpsdr.000035Patients with Triple-Negative Breast Cancer (TNBC), a highly heterogeneous and invasive subtype of breast cancer, do not benefit from hormonal therapy or trastuzumab; therefore, chemotherapy is considered the only option. We explored the effect of the chemotherapeutic drug cyclophosphamide (Cytoxan; Cy) on TNBC by an integrated bioinformatics approach.
Methods: Pharm Mapper, Gene Cards, and Swiss Target Prediction were used to identify potential targets of Cy. Differentially expressed genes (DEGs) in TNBC were screened out from four GEO datasets. Common genes were further evaluated by a protein-protein interaction network analysis, core gene identification, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses, docking assays, and survival and immune cell infiltration analyses.
Results: We collected information on 1638 drug targets and 494 DEGs in TNBC, including 267 up-regulated and 227 down-regulated genes. In total, 68 overlapping genes were identified as common targets. Ten core genes were identified in network analysis; GO and KEGG analyses revealed enrichment for DNA damage and many signaling pathways. Four core gene targets were verified by molecular docking. Kaplan–Meier analysis revealed that two core genes were significantly related to an adverse overall survival; furthermore, immune infiltration analysis suggested that Cy affects the microenvironment.
Conclusions: Our integrative bioinformatics approach revealed that the anti-TNBC effect of Cy was mediated by DNA damage-related genes and many pathways. These findings provide a basis for further functional studies aimed at improving outcomes in TNBC.
Introduction
Triple-Negative Breast Cancer (TNBC) is a breast cancer subtype characterized by no or low expression of Estrogen Receptor (ER), Progesterone Receptor (PR), and Human Epidermal Growth Factor Receptor 2 (HER2) [1]. Compared with other breast cancer subtypes, the clinical-pathological features of TNBC usually include larger tumors, a worse histologic grade, and higher proliferation index [2]. with high heterogeneity and invasiveness. A study of National Cancer Institute Surveillance, Epidemiology and End Results (SEER) from 2010 to 2012 revealed that TNBC has a worse overall survival and cause-specific survival than non-TNBC [3]. The 5-year overall survival rate of TNBC is <78.5%, and there is a high risk of distant recurrence in the first 3 to 5 years following diagnosis [3,4].
Although the mechanisms underlying TNBC are not fully understood, approximately 10% of patients have inactivating mutations in BRCA1 or BRCA2 [5]. Transcriptomic profiling has shown that multiple genomic transformations activate the PI3K pathway, including the activation of neoplastic genes and inactivation of tumor suppressor genes, thus altering the PI3K signaling pathway [6]. PI3K changes at the genetic level result in aberrations in the AKT/mTOR pathway [7]. The Hedgehog pathway, which is a highly conserved and critical signaling pathway, plays an important role in regulating embryonic mammary gland induction and the ductal architecture and differentiation in lactation [7]. Increasing evidence suggests that upregulation of the Hh ligand sonic hedgehog is correlated with inferior overall survival in TNBC [8]. Epidermal Growth Factor Receptor (EGFR) is a growth indicator in tumors; in TNBC, the upregulation of EGFR is associated with tumor progression [9]. and is always coupled with high Vascular Endothelial Growth Factor (VEGF) expression, affecting the RAS–MAPK signaling pathway [10]. Many factors involved in the proliferation of TNBC have been reported, including AMP-Activated Protein Kinase (AMPK) [11]. Mouse Double Minute 2 Homolog (MDM2) [12] and Metadherin (MTDH) [13].
Cyclophosphamide (Cytoxan; Cy) has been a widely used chemotherapeutic agent for immunotherapy in various cancer types since it was first synthesized. It is usually combined with platinum-based drugs [14]. taxane-based drugs [15] and anthracycline-based drugs [16] as Neoadjuvant Chemotherapy (NACT) in TNBC. It has been reported to function by three distinct antitumor mechanisms, namely, the inhibition of nuclear DNA replication, prevention of DNA synthesis and RNA transcription from the affected DNA, and mispairing of nucleotides, leading to mutations.
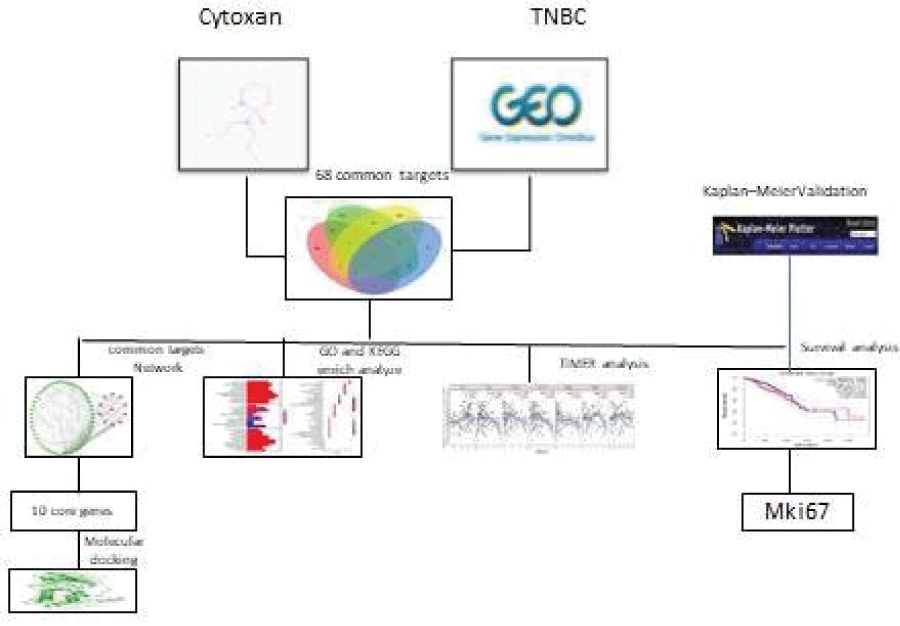
Cy often leads to chemo-toxicity; however, in some cases, it is the only available therapeutic choice to reduce and prevent tumor relapse. To reveal the mechanisms by which Cy exerts anti-TNBC effects, we used a network pharmacology approach to study its targets and related pathways in the treatment of TNBC, with validation by survival analyses. The workflow is shown in Figure 1.
Material and methods
Cy target information
Cy target information was extracted from Pharm Mapper (http://lilab- ecust.cn/pharmmapper/), GeneCards (https://www.genecards.org/), and Swiss Target Prediction (http://www.swisstargetprediction.ch/).
TNBC target information from GEO
Four gene expression datasets were downloaded from Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov). All datasets were separated into two groups (TNBC and non-TNBC) based on the original data, and the basal-like subtype was classified as TNBC [17]. Table 1 provides detailed information for the four GEO datasets.
Identification of differentially expressed genes
Using the R computing environment (R×64 3.5.3, https://cran.r-project.org/), a Robust Rank Aggregation (RRA) analysis was performed to evaluate the four datasets [18,19].
The limma package in R http://www.bioconductor.org/packages/release/bioc/html/limma.html) was used to identify differentially expressed genes (DEGs) between the TNBC and non-TNBC groups. Hub genes with |Log2FC| > 1 and adjusted-P < 0.05 were regarded as DEGs.
Overlap between TNBC-related DEGs from GEO and Cy targets
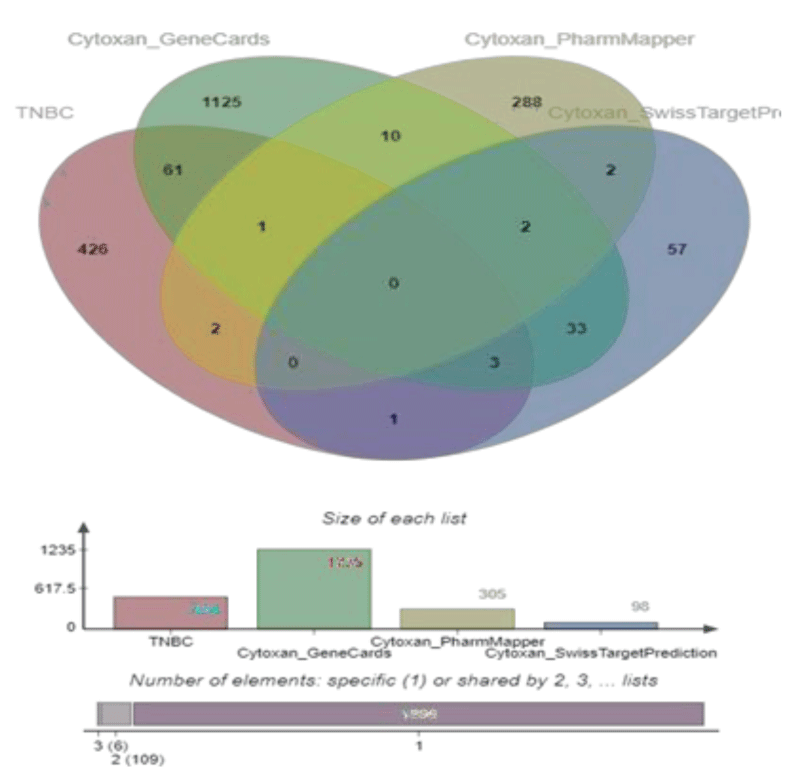
The DEGs in TNBC and potential targets of Cy were uploaded to E Venn (http://www.ehbio.com/test/venn/#/) to obtain 68 overlapping genes, which are visually displayed in a Venn diagram in Figure 2. All 68 overlapping genes were considered common targets and included in further analyses.
Constructions of PPI networks
Protein-protein interaction (PPI) networks of common targets were constructed using STRING (https://string-db.org/), which includes protein interaction information covering a large number of taxa; Homo sapiens was selected as the study object. Then, the STRING network results were exported to Cytoscape (version 3.8.2, https://cytoscape.org/), The MCODE package was used to visualize and identify core genes in PPI networks based on the following criteria: MCODE score >5 and number of nodes >20.
GO and KEGG enrichment analyses
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses of core genes were performed using "ClusterProfiler” (http://bioconductor.org/pages/release/bioc/html/clusterProfiler.html) in R.
Molecular docking
Molecular docking was used to explore the binding interactions between drugs and small molecule compounds, such as proteins. The structure of Cy was transformed into a three-dimensional structure using Chem3D Ultra 14.0 (CambridgeSoft Corporation, Cambridge, MA, USA). The MM2 method was used for energy minimization, and results were saved in mol format and exported as a pdbqt ligand parameter file using Autodocktools-1.5.6.
The protein structures UBE2C (PDB ID: 1i7k), CHEK1 (PDB ID: 2e9u), MKI67 (PDB ID: 1r21), TYMS (PDB ID: 6qyq), CCNA2 (PDB ID: 1oi9), BIRC5 (PDB ID: 3ued), TTK (PDB ID: 3gfw), CENPF (PDB ID: 6m4i), PLK (PDB ID: 2ogq), EZH2 (PDB ID: 4mi5) of core genes were evaluated using MGLTools v1.5.6, including hydrogenation, circuit calculations, and merging nonpolar hydrogens. All data were exported as pdb files and saved in pdbqt format.
Autodock Vina 1.1.2 was used for molecular docking; the conformation with the highest affinity was chosen as the final docking conformation, and results were visualized using Maestro 11.9.
Associations between core genes and survival and immune infiltration Kaplan–Meier plotter (http://kmplot.com/analysis/) was used to determine the influence of core genes on prognosis [20]. The data sources included GEO, EGA, and TCGA. The primary purpose of the tool is meta-analysis-based discovery and validation of survival biomarkers.
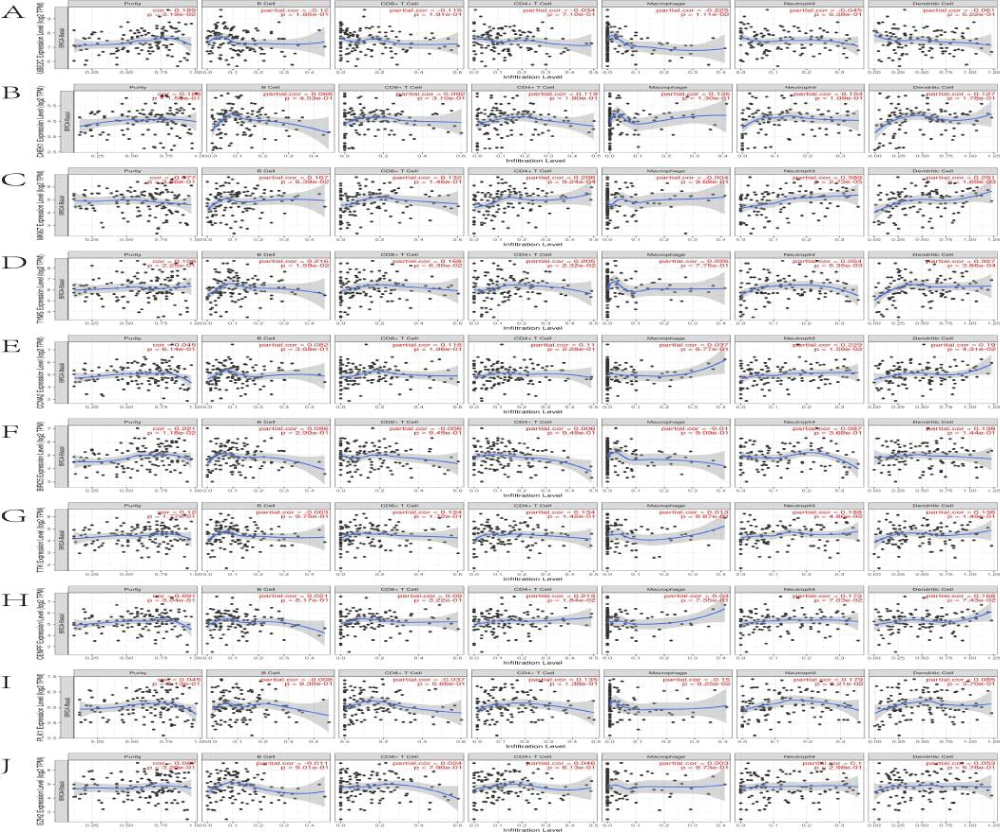
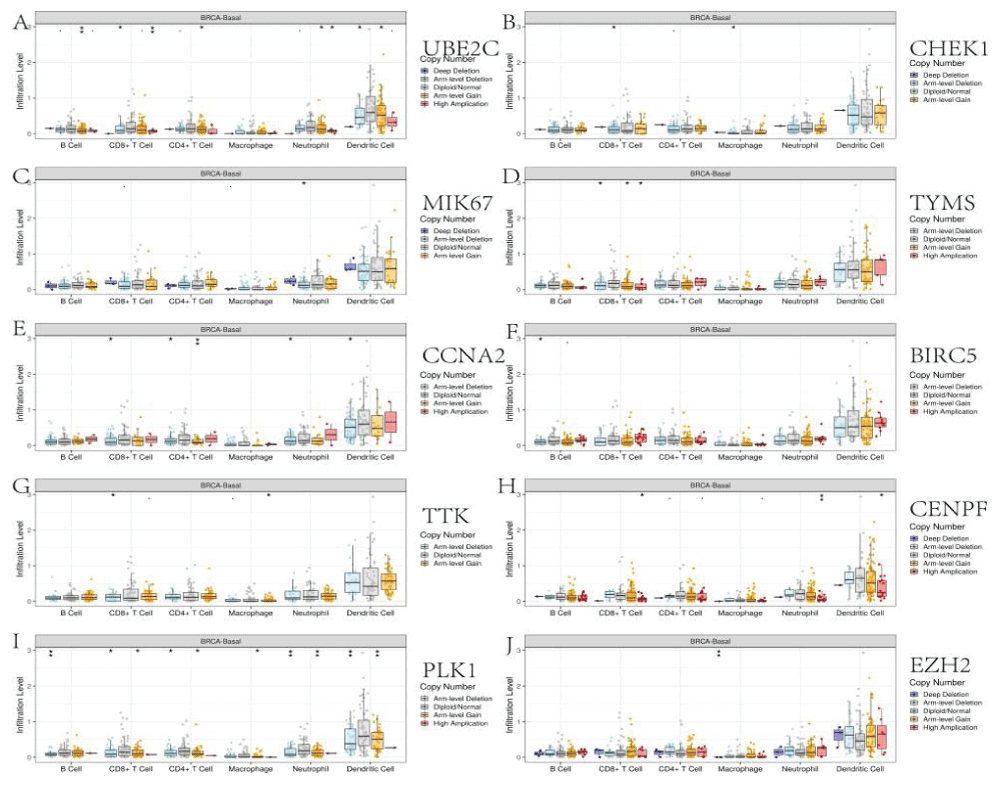
The Tumor Immune Estimation Resource database (TIMER, https://cistrome.shinyapps.io/timer/) applies a deconvolution method to infer the abundance of tumor-infiltrating immune cells from gene expression profiles. We used TIMER to analyze the characteristics of ten core genes that were closely related to the anti-TNBC effects of Cy. Correlations between expression levels of ten core genes and the abundance of immune cells were evaluated by Spearman's correlation coefficients, setting the cancer type to BRCA (Breast Invasive Carcinoma)-Basal. The infiltration level for each Somatic Copy Number Alteration (SCNA) category was compared with the normal level by a two-sided Wilcoxon rank-sum test.
Results
Collection of Cy-related drug target information
We obtained 305 drug targets from Pharm Mapper, 1235 drug targets from Gene Cards, and 98 drug targets from Swiss Target Prediction.
Identification of DEGs in TNBC based on GEO datasets
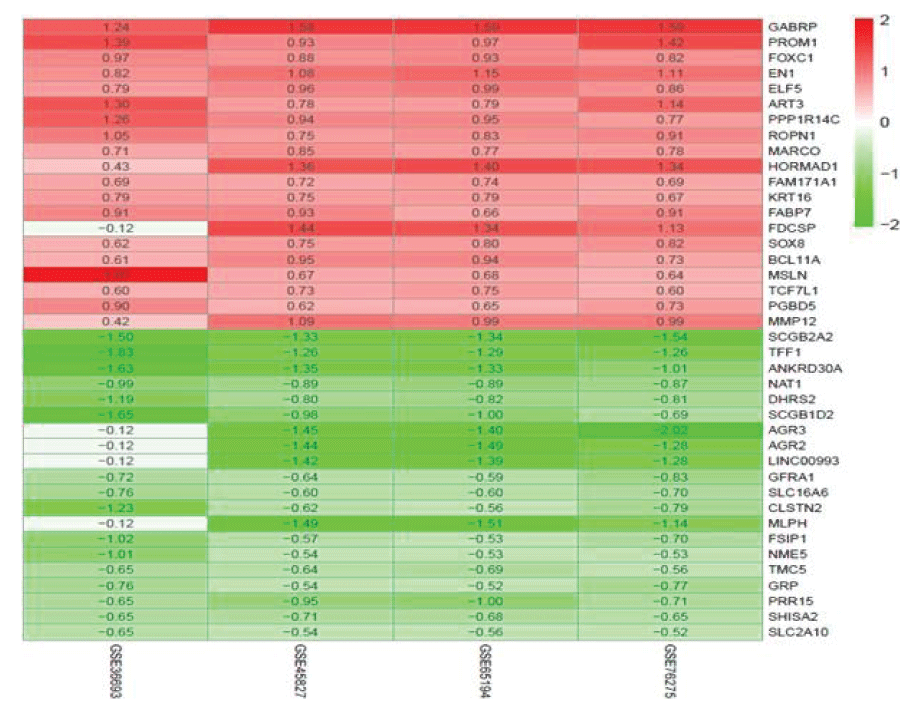
The RRA method was used for background correction and normalization. According to the criterion of 1.3 items, 494 DEGs were screened out, including 267 up-regulated and 227 down-regulated genes in TNBC. Figure 3 shows the top 20 up-regulated and down-regulated genes in the four datasets from GEO.
Construction of PPI networks
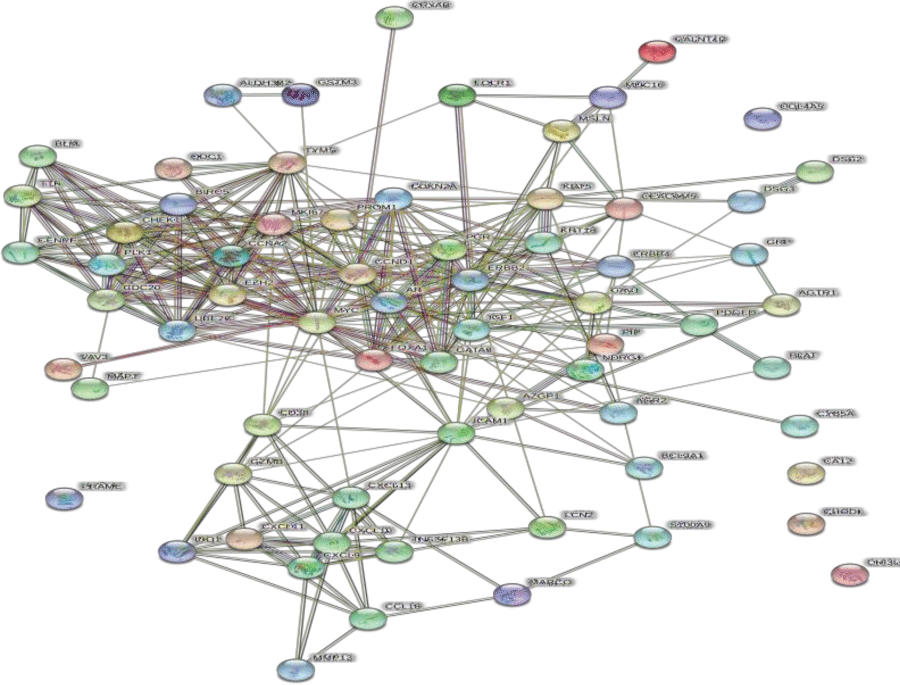
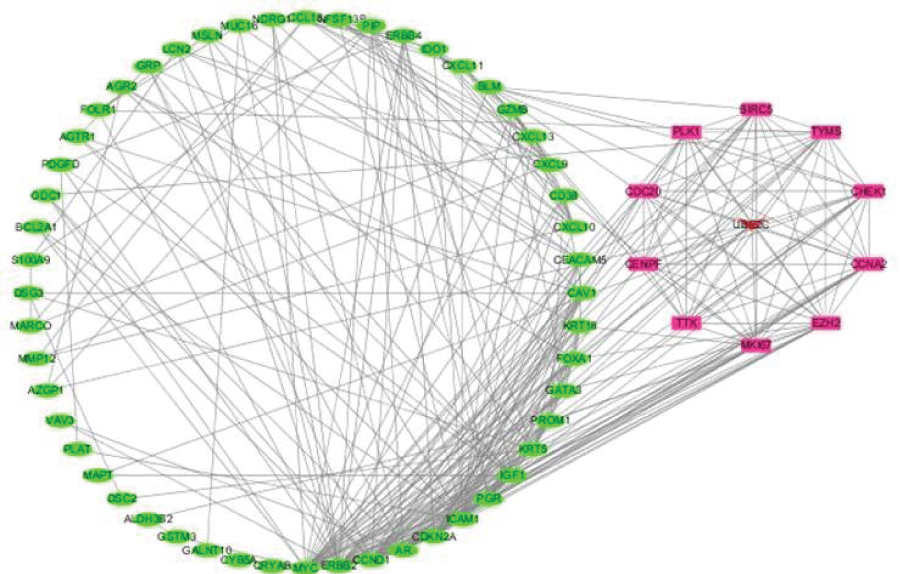
We constructed a PPI network with 68 nodes (based on the overlap between drug targets and DEGs) and 311 edges using STRING (Figure 4). The average node degree was 9.15, the average local clustering coefficient was 0.539, and the PPI enrichment p-value was <1.0e-16. Ten core genes were screened out using Cytoscape, including UBE2C, CHEK1, MKI67, TYMS, CCNA2, BIRC5, TTK, CENPF , PLK1, and EZH2 (Figure 5).
Functional enrichment analyses of candidate genes
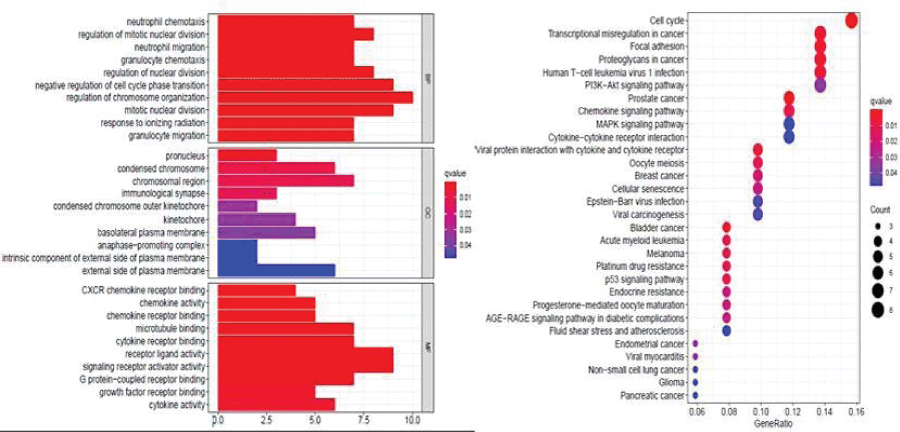
We evaluated all 68 targets of Cy with GO and KEGG pathway enrichment analyses and had the following key findings (Figure 6):
- The anti-TNBC effect of Cy involved the following biological processes: neutrophil chemotaxis, regulation of mitotic nuclear division, neutrophil migration, granulocyte chemotaxis, regulation of nuclear division, negative regulation of cell cycle phase transition, regulation of chromosome organization, mitotic nuclear division, response to ionizing radiation, and granulocyte migration.
- The anti-TNBC effect of Cy involved the following cellular components: pronucleus, condensed chromosome, chromosomal region, immunological synapse, condensed chromosome outer kinetochore, kinetochore, basolateral plasma membrane, anaphase−promoting complex, an intrinsic component of the external side of the plasma membrane, and the external side of the plasma membrane.
- The anti-TNBC effect of Cy involved the following molecular functions: CXCR chemokine receptor binding, chemokine activity, chemokine receptor binding, microtubule-binding, cytokine receptor binding, cytokine receptor binding, signaling receptor activator activity, G protein−coupled receptor binding, growth factor receptor binding, cytokine activity.
- KEGG enrichment analysis revealed four signaling pathways related to the anti- TNBC effects of Cy, including the p53 signaling pathway, PI3K−Akt signaling pathway, MAPK signaling pathway, and chemokine signaling pathway.
Molecular docking results
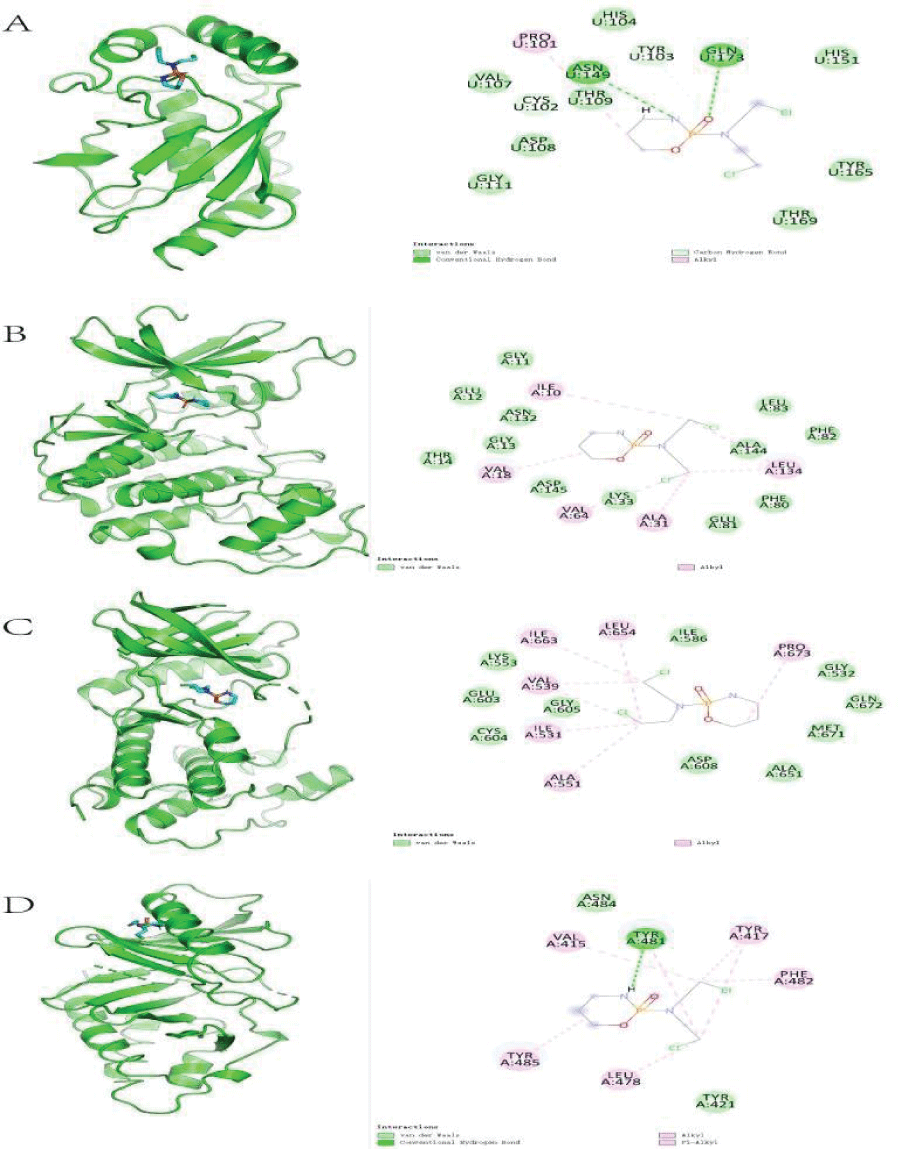
Molecular docking results revealed the 2-dimensional and 3-dimensional docking poses of Cy with each core target. Only four core targets showed successful docking with Cy (Figure 7), indicating that Cy may act via sites in these core targets.
Validation of core genes
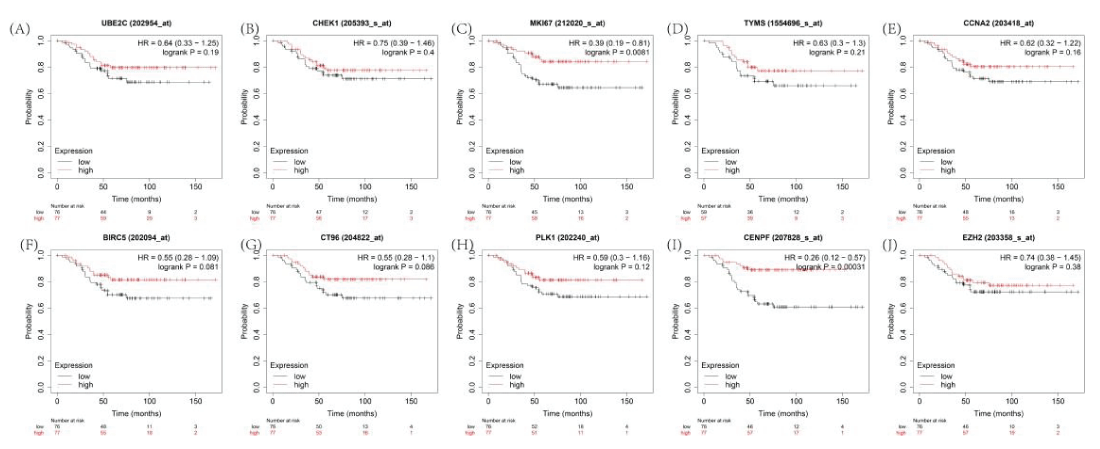
Based on a Kaplan–Meier analysis, 2 of 10 core genes were significantly related to an adverse overall survival in Cy-treated TNBC (Figure 8), including MKI67 (HR = 0.39 (0.19–0.18), P = 0.0081) and CENPF (HR = 0.26 (0.12−0.57), P = 0.00031).
There is documented evidence that MKI67 is a suitable genetic marker in TNBC; accordingly, we selected MKI67 as an efficient indicator of the efficacy of Cy against TNBC.
We explored the correlations between the levels of the 10 core genes and immune infiltration to evaluate the effects of Cy on the tumor microenvironment. Our results indicated that the core gene expression levels had a highly complex and variable effect on immune cell infiltration, reflecting the heterogeneity and complexity of the immune microenvironment in TNBC (Figure 9). The infiltration level for each SCNA illustrated the association with mutations in core genes in BRCA-Basal. The distributions of core gene subsets for each copy number status in BRCA-Basal are shown in box plots in Figure 10.
Discussion
TNBC, a highly heterogeneous and invasive subtype of breast cancer, has a poor prognosis, despite huge clinical investment. The median overall survival is less than 40 months [21]. TNBC usually occurs in young women carrying a BRAC1 mutation, especially in African-American women [22. The aggressive character of TNBC often involves the lung and central nervous system metastasis, rather than bone metastasis. Many clinicopathologic features are closely associated with TNBC, including younger age, bigger tumor size, higher rate of node positivity, and higher parity of tumor [23]. In clinical practice, surgery, and radiotherapy are considered the first-line therapeutic strategy in both TNBC and non-TNBC. Many additional therapeutic strategies have been evaluated in TNBC, such as targeted strategies and immunotherapy; however, outcomes remain poor.
In this study, we analyzed four genome‑wide datasets downloaded from GEO (based on the same platform, GPL-570) and used an integrated RRA method to reduce batch effects. We identified a substantial number of significantly upregulated or downregulated genes, some of which were known biomarkers of TNBC, such as GABRP, a potential therapeutic target [24,25]. androgen receptor FOXC1, an important immunohistochemical marker for the classification of TNBC [26]. homeobox transcription factor EN1, a key BRD4-S coregulator, particularly in TNBC [27]. Trefoil factor 1 (TFF1), a potential prognostic biomarker with functional significance in breast cancers [28]. and NAT1, with an important role in TNBC [29]. The functions of the other DEGs and mechanisms of action are poorly documented and should be a focus of future research. Cy is a prodrug that is activated by biotransformation phase I enzymes. After administration, most Cy is transformed to 4-hydroxycyclophosphamide by hepatic cytochrome P450 (CYP) enzymes [30]. The major site of Cy action is the liver [31]. It may form both intra- and interstrand DNA cross-links and DNA-protein cross-links, both resulting in the inhibition of DNA replication and cell death by apoptosis [32] however, the mechanism is unclear. We utilized an integrative bioinformatics approach to explore the effect of Cy in TNBC.
UBE2C, CHEK1, MKI67, TYMS, CCNA2, BIRC5, TTK, CENPF , PLK1, and EZH2 were core genes associated with the anti-TNBC effect of Cy. UBE2C and TTK are drivers of TNBC tumorigenesis based on a study of sensitivity to paclitaxel [33]. CHEK1, a conserved serine/threonine-protein kinase (also known as CHK1), is a major factor in halting the cell cycle in response to DNA damage [34] Chk1 inhibition by miR-320c increases oxaliplatin responsiveness in TNBC [35]. Studies on TNBC cell lines have shown that Chk1 inhibition leads to decreased cell viability via mitotic cell death and confirmed that it is a new therapeutic target [36]. MKI67 (proliferation marker protein Ki-67) is expressed in nuclei during different phases of the cell cycle, except in the G0 quiescent state. A study of 2465 female patients with TNBC revealed that a Ki-67 cutoff of 30% can be used for the further classification of TNBC into two subtypes with different responses and prognoses [37] Thymidylate Synthase (TYMS) is a rate-limiting enzyme in thymidylate biosynthesis. Song, et al. [38] demonstrated that there are significant correlations between TYMS expression and clinical parameters and survival in breast cancer, suggesting that TYMS functions in the onset and progression of breast cancer. Baculoviral inhibitor of apoptosis repeat-containing 5 (BIRC5) plays a vital role in carcinogenesis by influencing cell division and proliferation and by inhibiting apoptosis. BIRC5 expression is higher in various breast cancer subtypes than in matched controls [39] Turne, et al. screened 1,363 drugs in patient-derived xenograft (PDX) models and found that BIRC5 is highly expressed in basal-like PDXs, cell lines, and patient tumors and is negatively associated with outcomes [40]. Polo-Like Kinase-1 (PLK1) is correlated with a poor prognosis in TNBC. PLK1 overexpression is related to Ki67 [41]. Based on the functions of the core genes, the anti-TNBC effect of Cy is related to DNA damage, involving multiple targets.
GO and KEGG pathway enrichment analyses revealed that the differentially expressed core genes are enriched for various biological processes, including the regulation of chromosome organization and negative regulation of cell cycle phase transition. They were also enriched for cellular components, including the pronucleus, condensed chromosome, and chromosomal region components with the ability to promote cell cycle progression and increase invasion [42]. In the molecular function category, we detected enrichment for CXCR chemokine receptors, which regulate breast tumor metastasis and angiogenesis. The results further suggested that regulation of the cell cycle and transcriptional dysregulation are vital processes in the effect of Cy against TNBC.
Molecular docking results demonstrated the interactions between Cy and core genes. Cy functioned via different locations of different core genes, potentially conferring different pharmacological activities. However, further in vivo and in vitro analyses of the pharmacodynamic are needed to determine the utility of this finding.
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
- Bose S (2015) Triple-negative breast carcinoma: morphologic and molecular subtypes. Adv Anat Pathol 22: 306–313. Link: https://bit.ly/3EtTsRG
- Denkert C, Liedtke C, Tutt A, von Minckwitz G (2016) Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet 2016: 2430-2442. Link: https://bit.ly/3pl8mFe
- Li X, Yang J, Peng L, Sahin AA, Huo L, et al. (2017) Triple-negative breast cancer has worse overall survival and cause- specific survival than non-triple-negative breast cancer. Breast Cancer Res Treat 161: 279-287. Link: https://bit.ly/3eo4fBW
 Foulkes WD, Smith IE, Reis-Filho JS (2010) Triple negative breast cancer. N Engl J Med 363: 1938-1948. Link: https://bit.ly/3mOE6RN
Foulkes WD, Smith IE, Reis-Filho JS (2010) Triple negative breast cancer. N Engl J Med 363: 1938-1948. Link: https://bit.ly/3mOE6RN- Tutt A, Tovey H, Cheang MCU , Kernaghan S, Kilburn L , et al. (2018) Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: the TNT Trial. Nat Med 24: 628-637. Link: https://bit.ly/3H3QZ1O
 Pascual J, Turner NC (2019) Targeting the PI3-kinase pathway in triple-negative breast cancer. Ann Oncol 30: 1051-1060. Link: https://bit.ly/3H1xwie
Pascual J, Turner NC (2019) Targeting the PI3-kinase pathway in triple-negative breast cancer. Ann Oncol 30: 1051-1060. Link: https://bit.ly/3H1xwie Bhateja P, Cherian M, Majumder S, Ramaswamy B (2019) The Hedgehog Signaling Pathway: A Viable Target in Breast Cancer?. Cancers 11: 1126. Link: https://bit.ly/3EzZpMZ
Bhateja P, Cherian M, Majumder S, Ramaswamy B (2019) The Hedgehog Signaling Pathway: A Viable Target in Breast Cancer?. Cancers 11: 1126. Link: https://bit.ly/3EzZpMZ- Noman AS, Uddin M, Rahman MZ, Nayeem MJ, Alam SS, et al. (2016) Overexpression of sonic hedgehog in the triple negative breast cancer: clinicopathological characteristics of high burden breast cancer patients from Bangladesh. Sci Rep 6: 18830. Link: https://bit.ly/32eLSgH
- Liu Y, Zhou Y, Huang KH, Li Y, Fang X, et al. (2019) EGFR-specific CAR-T cells trigger cell lysis in EGFR-positive TNBC. Aging (Albany NY) 11: 11054-11072. Link: https://bit.ly/3ezd4cB
- Giltnane JM, Balko JM (2014) Rationale for targeting the Ras/MAPK pathway in triple-negative breast cancer. Discov Med 17: 275-283. Link: https://bit.ly/32lBbZB
- Cao L, Niu Y (2020) Triple negative breast cancer: special histological types and emerging therapeutic methods. Cancer Biol Med 17: 293-306. Link: https://bit.ly/3EtTvgk
- Gao C, Xiao G, Piersigilli A, Gou J, Ogunwobi O, et al. (2019) Context-dependent roles of MDMX (MDM4) and MDM2 in breast cancer proliferation and circulating tumor cells. Breast Cancer Res 21: 5. Link: https://bit.ly/3yUl1T1

- Tan L, Qin H, Piao Y, Liu Z, Han Y, et al. (2015) Expression and clinical significance of MTDH and VEGF in triple-negative breast cancer. Zhonghua Zhong Liu Za Zhi 37: 827-832. Link: https://bit.ly/32iTI8M
- Nakatsukasa K, Koyama H, Oouchi Y, Imanishi S, Mizuta N, et al. (2016) Docetaxel and cyclophosphamide as neoadjuvant chemotherapy in HER2-negative primary breast cancer. Breast Cancer 24: 63-68. Link: https://bit.ly/3qhKHEZ
- Loibl S, Untch M, Burchardi N, Huober J, Sinn BV, et al. (2019) A randomised phase II study investigating durvalumab in addition to an anthracycline taxane-based neoadjuvant therapy in early triple-negative breast cancer: clinical results and biomarker analysis of GeparNuevo study. Ann Oncol 30: 1279-1288. Link: https://bit.ly/3qhdMAB
- Schneeweiss A, Chia S, Hickish T, Harvey V, Eniu A, et al. (2013) Pertuzumab plus trastuzumab in combination with standard neoadjuvant anthracycline-containing and anthracycline-free chemotherapy regimens in patients with HER2-positive early breast cancer: a randomized phase II cardiac safety study (TRYPHAENA). Ann Oncol 24: 2278-2284. Link: https://bit.ly/3yR7BH8
- Bose S (2015) Triple-negative breast carcinoma: morphologic and molecular subtypes. Adv Anat Pathol 22: 306-313. Link: https://bit.ly/3FsmKBi
- Kolde R, Laur S, Adler P, Vilo J (2012) Robust rank aggregation for gene list integration and meta- analysis. Bioinformatics 28: 573-580. Link: https://bit.ly/33SWhit
- Zhong G, Lou W, Shen Q, Yu K, Zheng Y (2020) Identification of key genes as potential biomarkers for triplenegative breast cancer using integrating genomics analysis. Mol Med Rep 21: 557566. Link: https://bit.ly/3ms9pl3
- Nagy A, Munkárcsy G, Győrffy B (2021) Pancancer survival analysis of cancer hallmark genes. Sci Rep 11: 6047. Link: https://bit.ly/3Jdg9wV
- Gluz O, Liedtke C, Gottschalk N, Pusztai L, Nitz U, et al. (2009) Triple-negative breast cancer--current status and future directions. Ann Oncol 20: 1913–1927. Link: https://bit.ly/32pPpbM
- Diaz LK, Cryns VL, Symmans WF, Sneige N (2007) Triple negative breast carcinoma and the basal phenotype: from expression profiling to clinical practice. Adv Anat Pathol 14: 419-430. Link: https://bit.ly/3H6c77M
- Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, et al. (2006) Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA 295: 2492-2502. Link: https://bit.ly/3yQCXOl
- Wali VB, Patwardhan GA, Pelekanou V, Karn T, Cao J, et al. (2019) Identification and Validation of a Novel Biologics Target in Triple Negative Breast Cancer. Sci Rep 9: 14934. Link: https://go.nature.com/3suvyCT
- Li X, Wang H, Yang X, Wang X, Zhao L, et al. (2021) GABRP sustains the stemness of triple-negative breast cancer cells through EGFR signaling. Cancer Lett 514: 90-102. Link: https://bit.ly/3qmpERD
- Zhao S, Ma D, Xiao Y, Li XM, Ma JL, et al. (2020) Molecular Subtyping of Triple-Negative Breast Cancers by Immunohistochemistry: Molecular Basis and Clinical Relevance. Oncologist 25: e1481-e1491. Link: https://bit.ly/3egA0gn
- Wu SY, Lee CF, Lai HT, Yu CT, Lee JE, et al. (2020) Opposing Functions of BRD4 Isoforms in Breast Cancer. Mol Cell 78: 1114-1132.e10. Link: https://bit.ly/3sqveFq
- Yi J, Ren L, Li D, Wu J, Li W, et al. (2020) Trefoil factor 1 (TFF1) is a potential prognostic biomarker with functional significance in breast cancers. Biomed Pharmacother 124: 109827. Link: https://bit.ly/3stZvTH
- Fei H, Chen S, Xu C (2020) RNA-sequencing and microarray data mining revealing: the aberrantly expressed mRNAs were related with a poor outcome in the triple negative breast cancer patients. Ann Transl Med 8: 363. Link: https://bit.ly/3FnoBal
- Hajdinák P, Szabó M, Kiss E, Veress L, Wunderlich L, et al. (2020) Genetic Polymorphism of GSTP-1 Affects Cyclophosphamide Treatment of Autoimmune Diseases. Molecules 25: 1542. Link: https://bit.ly/3solVG2
- de Jonge ME, Huitema AD, Rodenhuis S, Beijnen JH (2005) Clinical pharmacokinetics of cyclophosphamide. Clin Pharmacokinet 44: 1135-1164. Link: https://bit.ly/3ySLRdX
- Shulman-Roskes EM, Noe DA, Gamcsik MP, Marlow AL, Hilton J, et al. (1998) The partitioning of phosphoramide mustard and its aziridinium ions among alkylation and P-N bond hydrolysis reactions. J Med Chem 41: 515-29. Link: https://bit.ly/3yQ7Ylr
- Elango R, Vishnubalaji R, Shaath H, Alajez NM (2021) Molecular subtyping and functional validation of TTK, TPX2, UBE2C, and LRP8 in sensitivity of TNBC to paclitaxel. Mol Ther Methods Clin Dev 20: 601-614. Link: https://bit.ly/3mtGmh0
- Liu XP, Huang Q, Yin XH, Meng XY, Cao Y, et al. (2020) Strong Correlation between the Expression of CHEK1 and Clinicopathological Features of Patients with Multiple Myeloma. Crit Rev Eukaryot Gene Expr 30: 349-357. Link: https://bit.ly/3qi6IDL
- Lim S, Kim Y, Lee SB, Kang HG, Kim DH, et al. (2020) Inhibition of Chk1 by miR-320c increases oxaliplatin responsiveness in triple-negative breast cancer. Oncogenesis 9: 91. Link: https://go.nature.com/3pmYB9H
- Albiges L, Goubar A, Scott V, Vicier C, Lefèbvre C, et al. (2014) Chk1 as a new therapeutic target in triple- negative breast cancer. Breast 23: 250-258. Link: https://bit.ly/3yV38n6
- Zhu X, Chen L, Huang B, Wang Y, Ji l, et al. (2020) The prognostic and predictive potential of Ki-67 in triple- negative breast cancer. Sci Rep 10: 225. Link: https://go.nature.com/3HnjUyf
- Song S, Tian B, Zhang M, Gao X, Jie L, et al. (2021) Diagnostic and prognostic value of thymidylate synthase expression in breast cancer. Clin Exp Pharmacol Physiol 48: 279-287. Link: https://bit.ly/3HawujU
- Turner TH, Alzubi MA, Harrell JC (2020) Identification of synergistic drug combinations using breast cancer patient-derived xenografts. Sci Rep 10: 1493. Link: https://go.nature.com/3yQPWQ2
- Turner TH, Alzubi MA, Harrell JC (2020) Identification of synergistic drug combinations using breast cancer patient-derived xenografts. Sci Rep 10: 1493. Link: https://go.nature.com/3yTZkSX
- Ueda A, Oikawa K, Fujita K, Ishikawa A, Sato E, et al. (2019) Therapeutic potential of PLK1 inhibition in triple-negative breast cancer. Lab Invest 99: 1275-1286. Link: https://go.nature.com/3Epvk2p
- Zhang Y, Liu F, Zhang C, Ren M, Kuang M, et al. (2019) Non-SMC Condensin I Complex Subunit D2 Is a Prognostic Factor in Triple-Negative Breast Cancer for the Ability to Promote Cell Cycle and Enhance Invasion. Am J Pathol 190: 37-47. Link: https://bit.ly/3ektx48
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
