International Journal of Pharmaceutical Sciences and Developmental Research
An overview on NMR spectroscopy based metabolomics
Nalini Kanta Sahoo*, G Tejaswini, Madhusmita Sahu and KS Muralikrishna
Cite this as
Sahoo NK, Tejaswini G, Sahu M, Muralikrishna KS (2020) An overview on NMR spectroscopy based metabolomics. Int J Pharm Sci Dev Res 6(1): 016-020. DOI: 10.17352/ijpsdr.000029Introduction: A field of bioscience analysis that uses High throughput (HT) technologies to identify the products of metabolism formed in the cell metabolic processes (i.e. the metabolome). This process is termed metabolomics.The first NMR based metabolic studies were carried out by Wilson and Burlingame in the year 1974. Nuclear magnetic spectroscopy is one technique using which one can identify and characterize the metabolites formed due to metabolism.
Methodology: The NMR workflow is as follows:
Sample preparation ➨ sample loading and spectral acquisition ➨ spectral processing ➨ Spectral deconvolution ➨ data interpretation.
Conclusion: In the upcoming years, it is expected that more works will be reported depending upon the advantages and strengths that the NMR spectroscopy offers with regard to metabolomics. The NMR spectroscopy technique offers high versatility, and specificity with respect to spectra. This technique will gain high potential in the future because of its reproducibility and the ability to quantify the metabolites.
Introduction
Metabolomics
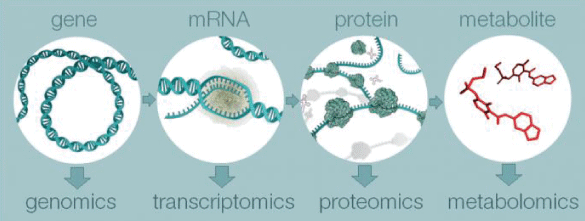
Metabolomics is defined as the comprehensive analysis of metabolites i.e., identification and quantification in a biological system Figure 1 [1].
The metabolites known by metabolomics square measure intermediary metabolites used to form the macromolecular structures and some other molecules that help in necessary metabolic functions of the body. Also metabolites those are involved in signalling can also be identified along with the secondary metabolites [2].
The study of molecules whose molecular size is less than or equal to 1500 Daltons is called as metabolomics. The metabolomics studies do not involve the quantification of sugars, polymers and amino acids since their molecular weight will be more than 1500 Daltons [3].
The development of metabolomic research has allowed its application in different fields, such as in nutrition [4,5], the environment [6,7], chemotaxonomy [8,9], pharmacology, in discovering biomarkers for a given disease [10,11], etc.
Approaches for the analysis of metabolome
There are three approaches reported to identify and qualify the proteins. They are described as follows.
1. Metabolite profiling
2. Metabolic fingerprinting
3. Metabonomics
Metabolite profiling
The metabolite profiling is a technique which enables is said to be one of the major tools that helps to identify large number of metabolites. But this process is done on a biased scale because of the limitations involved in the methodology and differences in analytical platforms. This is one limitation for metabolite profiling. Despite the bias introduced, this method is still used widely for the analysis of metabolites that enables to give true and complete picture of the metabolome.
Metabolite fingerprinting
The high throughput technologies like NMR spectroscopy are used to identify and characterise the structure of metabolome. Just like every person has a unique fingerprint, similarly, every metabolite has unique characteristic comparable to the fingerprint of the human being. Comparison of these metabolites with change in time can be identified using the metabolite fingerprinting. This technique is relatively simple and coarser compared to metabolite profiling.
Metabonomics
Metabonomics is the study of biological and scientific processes that involve the metabolites. The Metabonomics mainly focuses on the metabolic response that the organism produces in response to the pathophysiological stimuli or due to the genetic modifications occurring in the body. Generally, the study of Metabonomics is restricted to analysis of metabolites in microbiological and non-botanical studies [12].
Although, these three different approaches generate the copious amounts of productive information about the metabolite, they do not provide the comprehensive analysis of the metabolome. But, they allow the scientist to get an idea of the metabolic state of an organism at a particular period of time i.e. it provides a snapshot of the metabolic state. Metabolomics can lead to better understanding of the organization and functioning of the organism when used along with other omic studies like genomics, proteomics, and other physiological assays [12].
Applications
The metabolomic studies have found to have great potential in plant biology. They are used to study various parameters like accumulation of biomass, study of production of secondary metabolites in plants, and also stress resistance [13].
Thanatometabolomics- study of metabolite modulations after death i.e., forensic studies [14].
Importance
1. Biomass accumulation and resistance to sure environmental stressors area unit vital as plants area unit sought-after as a possible supply of different energy production like biofuel.[12].
2. Precision medicine
3. Diagnosis of disease with analysis of metabolite responsible for the disease.
4. The presence of adulterants in food and medicines can also be identified with the help of metabolomics.
5. To know the time of death of person and to help in Tissue transplantation [13].
Measurement of the metabolome
The two approaches used in designing the metabolomics studies are:
Targeted approach
The targeted approach is used when one wants to measure a particular set of metabolites.
Ex: the effect of drug on the specific enzyme is one type of targeted approach where the therapeutic modifications and genetic modifications occurring because of the drug metabolism are studied.
Untargeted approach
Untargeted approach can be used when one wants to identify and quantify metabolites as many as possible present in the biological sample. Various types of metabolites can be identified using the untargeted approach.
The two techniques that are used to study the metabolic state in an organism are:
✓ Mass spectrometry (MS) based metabolomics.
✓ Nuclear magnetic resonance (NMR) spectroscopy based metabolomics.
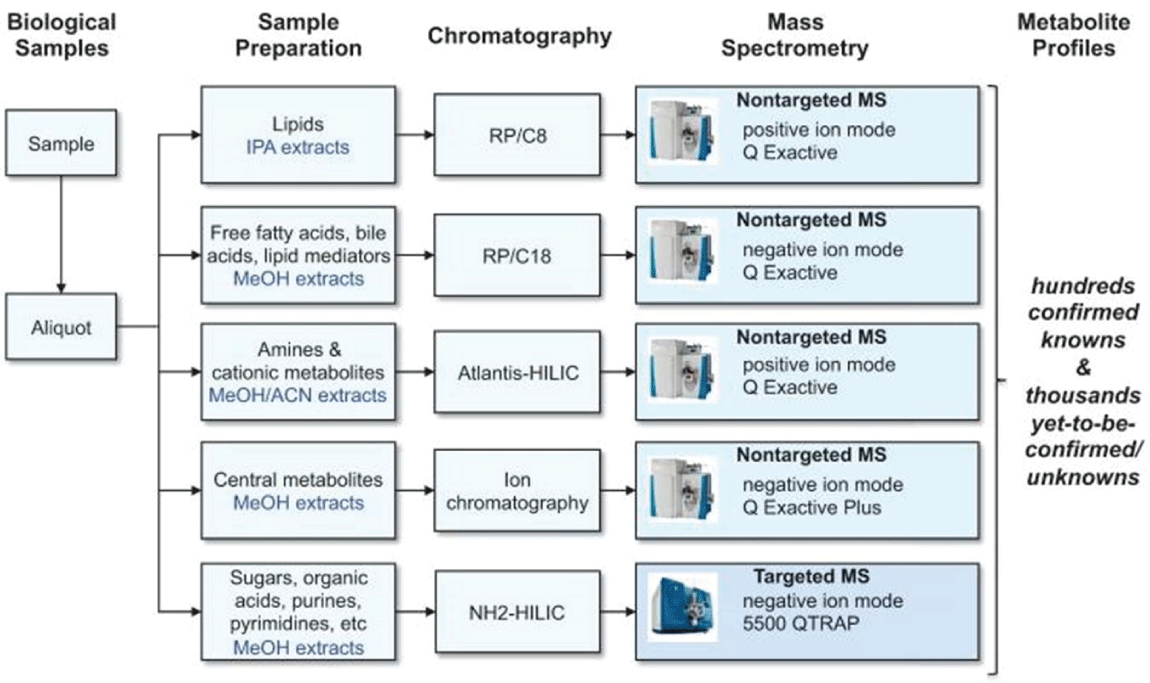
The ideal technique for metabolomic studies is the coupling of Mass spectroscopy with NMR spectroscopy. Though this process is quite expensive, better results can be expected Figure 2 [3].
Mass spectroscopy based metabolomics
The mass spectroscopy basically studies the effect of ionising energy on molecules. The principle involved in the technique of mass spectroscopy is as follows.
A mass spectrometer generated multiple ions form the sample that is under investigation. Then, electron ionisation occurs where the gas phase ion of the compund is produced. These molecules then undergo fragementation. Ions are then separated according to their mass to charge ratio and are detected. A mass spectrum is obtained, which is displayed in the form of plot of ion abundance versus mass to charge ratio. The study of ions reveal the information about the nature and structure of the precursor molecule. Also, molecular mass of the compund can be identified when the spectrum of the pure compund appears at highest value of mass to charge value Figure 3 [12].
NMR Spectroscopy
The nuclear magnetic resonance spectroscopy is a spectroscopic technique is used to determine the organic compounds. The molecular structure of the metabolite can be elucidated using the NMR spectroscopy.
Principle involved in NMR spectroscopy
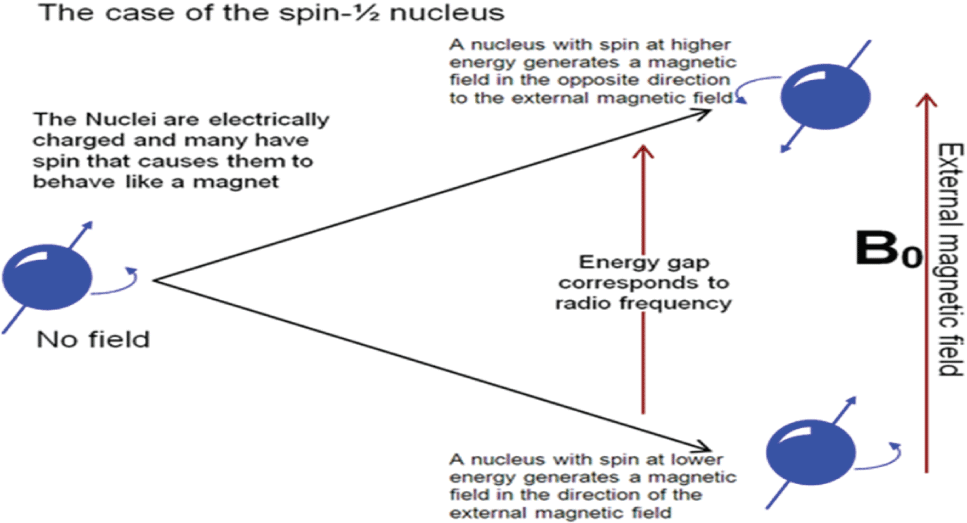
• All the metabolites are made up of atoms that consists of nuclei. Every nucleus consists of positive charges that is responsible for the spin of the nuclei.
• Whenever an external magnetic field is applied using any source, the atoms absorb the energy and excitation occurs. The electrons excite into the higher energy levels i.e. the energy transfer occurs at a particular wavelength.
• The wavelength at which energy transfer occurs corresponds to the frequency.
• When the external magnetic field is removed, the atoms come back to their original state into the lower energy levels. During this process, they emit energy at a particular frequency.
• The NMR spectrum of that particular nucleus is obtained for these energy transfers and frequencies [15].
The NMR spectrum gives the following information about the metabolite:
1. Number of signals produced by protons.
2. Position of the signals.
3. Integration and intensity of the signals.
4. Splitting of the signals produced.
The molecular structure of the metabolite can be determined from the H-NMR spectrum. The H-NMR can be utilized to put the complete structure of the metabolite together using the components of the spectra like signals, chemical shift, and integration and splitting patterns. The C-NMR spectroscopy abolishes some structures that can be possible if we use H-NMR spectrum Figure 4 [14].
History of NMR spectroscopy based metabolomics studies
• 1974 – first NMR based metabolic studies (Wilson and Burlingame)
• 1977-1984 – extensive C13 and p31 NMR studies of cell systems (Shulman)
• 1984 – first NMR characterization of human urine (Nicholson)
• 1986 – the “Fossel” NMR blood test for cancer is described (ET Fossel)
• 1991 – NMR based HDL/LDL test developed (Otvos)
• 1993 – LC model software developed for in vivo NMR (Provencher)
• 1999 – the word “ Metabonomics” was coined (Nicolson)
• 2000 – the word “Metabolomics” was coined (Drysdale)
• 2006-2016 – large metabolomic projects (Wilhart, Markley)
The workflow pattern of NMR based metabolomics
The workflow pattern can be categorized into 3 steps namely;
Sample preparation
The biological fluids like Cerebrospinal fluid require less or no preparation for NMR spectroscopy. Other biological fluids like plasma require the process of sample preparation. Because the plasma consists of proteins and lipids that may interfere with the quality of NMR spectrum. To remove the lipoproteins present in the plasma, methanol is used at solvent to serum ratio of 2:1. This ensures to minimize the loss of metabolites, and eliminating the undesired proteins and lipids.
Silica nanoparticles can also be used to remove proteins in the plasma [16].
Tagging
Tagging of compound with NMR active labels to is one best approach for compounds that show overlapping of 1H signals or those that are present in very less concentrations. Tagging with an NMR active label are useful in identification of the metabolite.
Nitrogen-15 with a hydrogen attached us the most widely used active label because of the advantage that it provided high sensitivity spectrum without the interference of other signals.
Quantification
For well resolved H-NMR peaks, the intensities of the signals linearly correlate with the relative concentration of the sample. To determine the concentration. Standard of known concentration should be added.
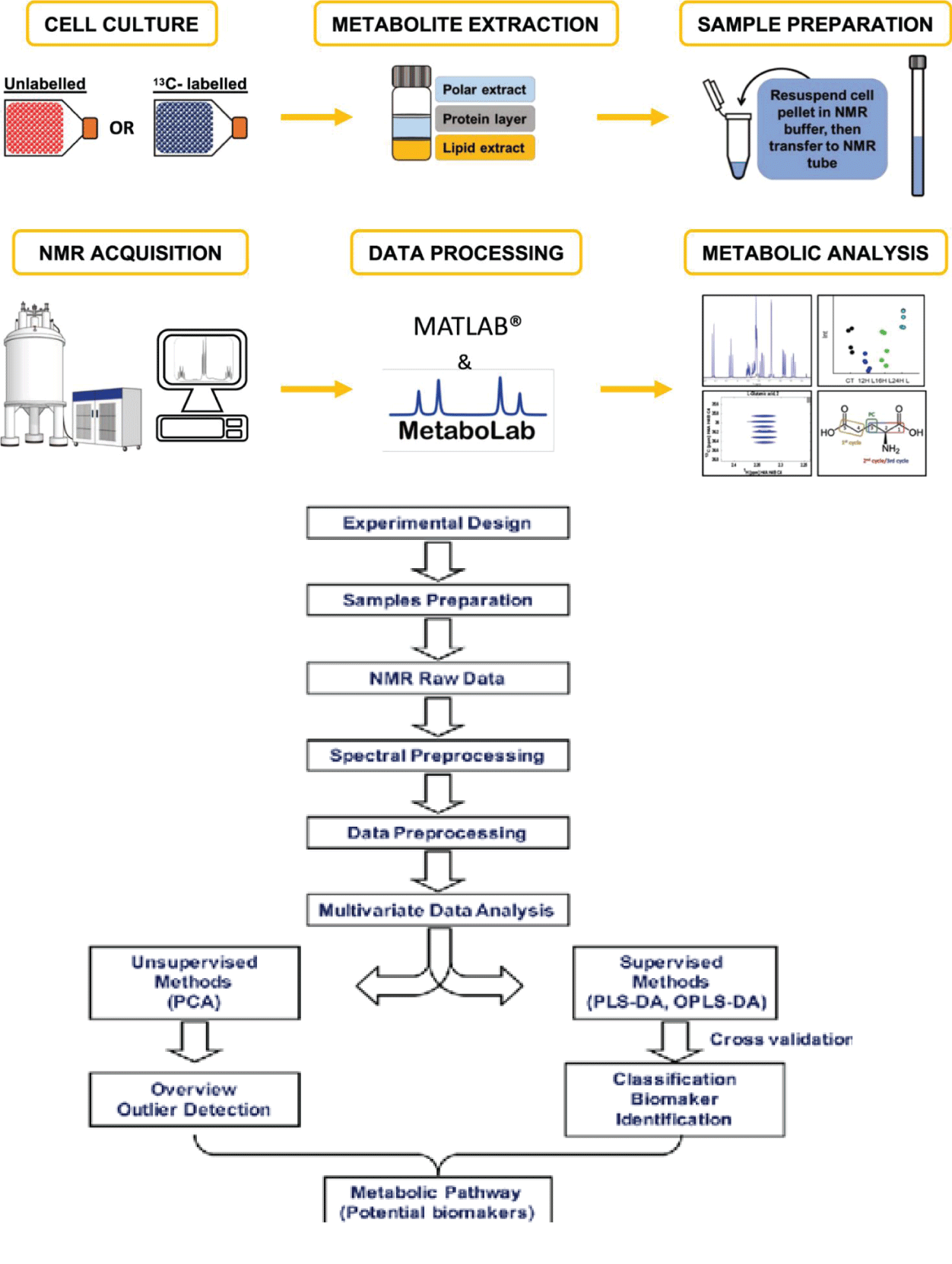
If the cross peak intensities of the same 2 dimensional H-C spectrum do not correlate with concentration, then one can collect the spectra of mixtures with the known concentration with known concentration bracketing those of unknown concentration. This is used to determine the factors that translate the maximum intensity into concentration. The peak intensities can also be converted into concentration by using the slopes that are generated by spectra utilizing various replicated Figure 5 [3].
Applications of NMR based metabolomics
1. Diagnosis of diseased condition.
2. Supervision of presence of adulterants in food and medicines, drugs.
3. Estimation of co-enzymes responsible for performing functions of the cell and quantification of those co-enzymes.
4. To find out changes in the redox potentials when exposed to toxins form external source of from within the cell.
5. Accelerating the new drug discovery process by identifying the lead compounds. The potency and toxicity of the lead compound that is known to elicit the pharmacological action are identified.
6. Removal of compounds that are known to cause certain problems and adverse effects in the body.
7. Identification of biomarkers in various cancers Figure 6 [3].
Future considerations
Though the method is fully developed, there is a need to concentrate more on developing the suitable databases for NMR data. Integrated and valid databases have to be developed in order to yield better results.
Suitable procedures of preparation of sample, NMR data collection, NMR data analysis should be suggested to enhance the accuracy of the results [2].
Advantages
1. NMR is ideal when the compounds are not possible to be ionized using the mass spectroscopy method.
2. Can also be used to identify those compounds that have identical masses.
3. For determining structures of unknown compounds.
4. Used to determine the mechanisms of the changes that the metabolite undergoes.
5. In experiments of screening of activity of drug [3].
Conclusion
In the upcoming years, it is expected that more works will be reported depending upon the advantages and strengths that the NMR spectroscopy offers with regard to metabolomics. The NMR spectroscopy technique offers high versatility, and specificity with respect to spectra. This technique will gain high potential in the future because of its reproducibility and the ability to quantify the metabolites.
- Clish CB (2015) Metabolomics: an emerging but powerful tool for precision medicine. Cold Spring Harb Mol Case Stud 1: a000588. Link: https://bit.ly/3gOX3is
- Barchet G A brief overview of metabolomics: what it means, how it is measured, and its utilization by Genoa Barchet. Link: https://bit.ly/2XYEPCz
- Markley JL, Brüschweiler R, Edison AS, Eghbalnia HR, Powers R (2017) The future of NMR-based metabolomics. Curr Opin Biotechnol 43: 34-40. Link: https://bit.ly/2Xtp7QK
- Hall RD, Brouwer ID, Fitzgerald MA (2008) Plant metabolomics and its potential application for human nutrition. Physiol Plant 132: 162–175. Link: https://bit.ly/3eKOlji
- Liu H, Tayyari F, Khoo C, Gu L (2015) A 1 H NMR-based approach to investigate metabolomic differences in the plasma and urine of young women aer cranberry juice or apple juice consumption. J Funct Foods 14: 76–86. Link: https://bit.ly/3coaEK1
- Lankadurai BP, Nagato EG, Simpson M (2013) Environmental metabolomics: an emerging approach to study organism responses to environmental stressors. Environ Rev 21: 180–205. Link: https://bit.ly/2MltC9E
- Soule MCK, Longnecker K, Jhonson WM, Kujawinski EB (2015) Environmental metabolomics: Analytical strategies. Mar Chem 177: 374-387. Link: https://bit.ly/2U4xbFk
- Kim HK, Saifullah S, Khan S, Wilson EG, Kricun SD, et al. (2010) Metabolic classication of South American Ilex species by NMR based metabolomics. Phytochemistry 71: 773–784. Link: https://bit.ly/3gNELOC
- Kim W, Peever TL, Park JJ, Park CM, Gang DR, et al. (2016) Use of metabolomics for the chemotaxonomy of legume associated Ascochyta and allied genera. Sci Rep 6: 20192. Link: https://go.nature.com/3cxe39n
- Chagas-Paula DA, Oliveira TB, Zhang T, Edrada-Ebel R, Da Costa FB (2015) Prediction of anti-inflammatory plants and discovery of their biomarkers by machine learning algorithms and metabolomic studies. Planta Med 81: 450-458. Link: https://bit.ly/2U4xHDg
- Chan AW, Mercier P, Schiller D, Bailey R, Robbins S, et al. (2015) 1 H-NMR urinary metabolomic profling for diagnosis of gastric cancer. Br J Cancer 114: 59–62. Link: https://bit.ly/2BnCsRX
- Emwas AH, Luchinat C, Turano P, Tenori L, Roy R, et al. (2015) Standardizing the experimental conditions for using urine in NMR-based metabolomic studies with a particular focus on diagnostic studies: a review. Metabolomics 11: 872–894. Link: https://bit.ly/304n8Uz
- Fan TW, Lane AN (2016) Applications of NMR spectroscopy to systems biochemistry. Prog Nucl Magn Reson Spectrosc 92-93: 18-53. Link: https://bit.ly/305CNmG
- Mora-Ortiz M, Trichard M, Oregioni A, Claus SP (2019) Thanatometabolomics: introducing NMR-based metabolomics to identify metabolic biomarkers of the time of death. 15: 37. Link: https://bit.ly/3gReGxZ
- Dunn WB, Ellis DI (2005) Metabolomics: current analytical platforms and methodologies. Trends in Analytical Chemistry Dunn WB, Ellis DI 24: 285-294. Link: https://bit.ly/2XqpTht
- Hata K, Higashisaka K, Nagano K, Mukai Y, Kamada H, et al. (2014) Evaluation of silica nanoparticle binding to major human blood proteins. Nanoscale research letters 668: 1-7. Link: https://bit.ly/2zZpaef
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
