International Journal of Agricultural Science and Food Technology
Review on comparative genome mapping in crop improvement
Zewdu Asrat* and Mastewal Gojjama
Cite this as
Asrat Z, Gojjam M (2022) Review on comparative genome mapping in crop improvement. Int J Agric Sc Food Technol 8(3): 218-224. DOI: 10.17352/2455-815X.000167Copyright License
© 2022 Asrat Z, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Comparative genomics is the study of the similarities and differences in the structure and function of hereditary information across taxa. The objective of this study was to highlight the role of comparative mapping in crop improvement. Hence, the study encompasses comparative genomics over the past two decades, multiple investigations of many additional taxa have delivered two broad messages: multiple investigations of many additional taxa have delivered two broad messages: In most plants, the evolution of the small but essential portion of the genome that actually encodes the organism’s genes has proceeded relatively slowly; as a result, taxa that have been reproductively isolated for millions of years have retained recognizable intragenic DNA sequences as well as similar arrangements of genes along the chromosomes. A wide range of factors, such as ancient chromosomal or segmental duplications, mobility of DNA sequences, gene deletion, and localized rearrangements, has been superimposed on the relatively slow tempo of chromosomal evolution. Comparative genomics is the study of the similarities and differences in the structure and function of hereditary information across taxa. The objective of this study was to highlight the role of comparative mapping in crop improvement. Hence, the study encompasses comparative genomics over the past two decades, multiple investigations of many additional taxa have delivered two broad messages: multiple investigations of many additional taxa have delivered two broad messages: In most plants, the evolution of the small but essential portion of the genome that actually encodes the organism’s genes has proceeded relatively slowly; as a result, taxa that have been reproductively isolated for millions of years have retained recognizable intragenic DNA sequences as well as similar arrangements of genes along the chromosomes. A wide range of factors, such as ancient chromosomal or segmental duplications, mobility of DNA sequences, gene deletion, and localized rearrangements, has been superimposed on the relatively slow tempo of chromosomal evolution.
Introduction
In the past ten years, there was great progress in linking plant genomes through comparative genetic maps, especially for species belonging to the same family [1]. Genetic mapping employs methods for the identification of the locus of a gene as well as for the determination of the distance between two genes [2]. Gene mapping is considered the major area of research in which molecular markers are used today. The principle of genetic mapping is chromosomal recombination during meiosis which results in the segregation of genes [3]. Comparative mapping can identify inversions, translocation, and duplications that have occurred. Genetic factors can also be assessed by comparing map distances of genes with conserved gene order in the two species. Therefore, the objective of this paper is to review comparative genome mapping in crop improvement.
Foundations of comparative genomics
Comparative genomics, the study of the similarities and differences in structure and function of hereditary information across taxa, uses molecular tools to investigate many notions that long preceded the identification of DNA as the hereditary molecule. Vavilov’s [4] law of homologous series in variation was an early suggestion of the similarities in the genetic blueprints of many plant species. Genetic analysis based on morphological and isoenzyme markers hinted at parallel arrangements of genes along the chromosomes of various taxa. These hints were later borne out at the DNA level, in seminal investigations of nightshades [5]. Over the past two decades, multiple investigations of many additional taxa have delivered two broad messages: (1) In most plants, the evolution of the small but essential portion of the genome that actually encodes the organism’s genes has proceeded relatively slowly; as a result, taxa that have been reproductively isolated for millions of years have retained recognizable intragenic DNA sequences as well as similar arrangements of genes along the chromosomes. (2) A wide range of factors, such as ancient chromosomal or segmental duplications, mobility of DNA sequences, gene deletion, and localized rearrangements, has been superimposed on the relatively slow tempo of chromosomal evolution and causes many deviations from co-linearity.
Origins of genome mapping
Despite advances in creating new genomic tools, in some cases revisiting old approaches to scientific questions can be fruitful. This retrospective strategy brings to mind the title of a popular show tune, ‘Everything old is new again’ [6]. In the case of genome mapping, something old is indeed new again. For many years, cytogeneticists looked at banding patterns of condensed chromosomes and made significant deductions and contributions to our understanding of plant genome organization. In recent years, improved optics, advanced molecular biology, and creative innovations have been combined to create higher-throughput genomic tools that have roots in, and similarities to, many older cytogenetic methods. These strategies produce maps of large individual DNA molecules. One reason why these long-molecule maps are receiving attention is because of their ability to complement genome sequencing. The relative ease of genome sequencing often overshadows its shortcomings: a puzzle with many small pieces is difficult to solve without additional, long-range information.
For example, genome maps can be combined with sequence assemblies comprising numerous scaffolds and consign, in which case they provide the necessary structure for joining contigs and improving the de novo assembly of plant genomes. Aside from de novo genome assembly, genome mapping provides some unique research opportunities for comparative plant genomics, which were previously closed because short sequencing reads cannot detect certain large structural variations. Here, we review genome mapping, including its limitations and capacities, and explore some of its potential applications in the field of plant comparative genomics.
Comparing plant genomes
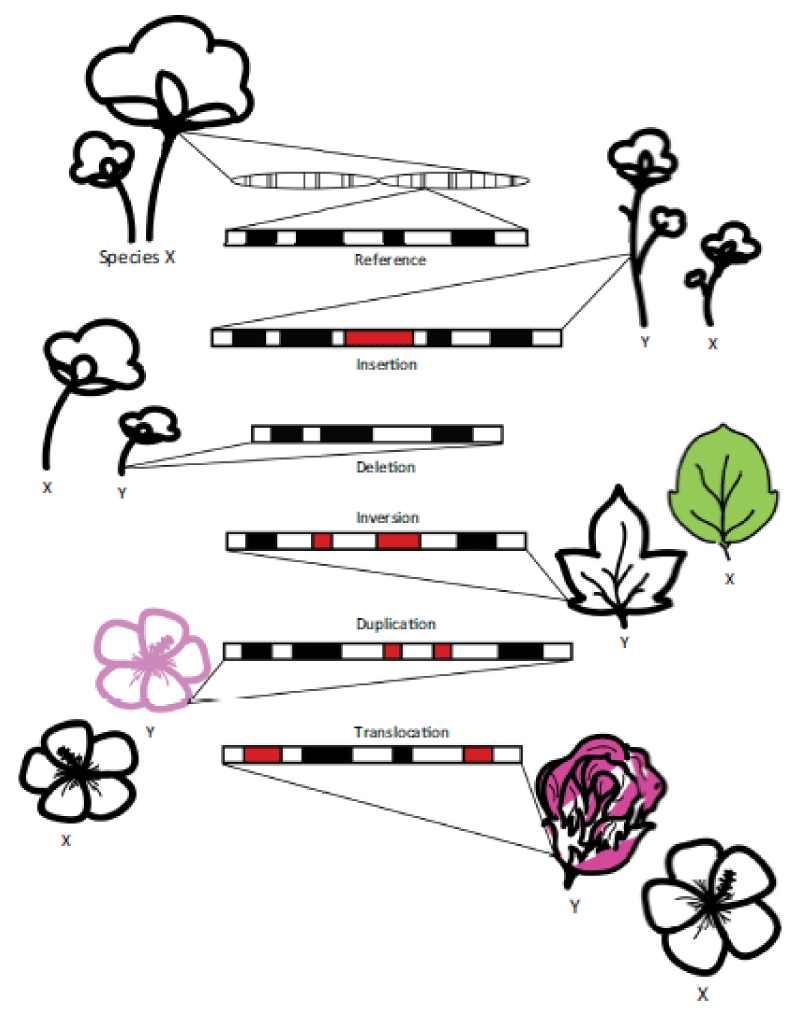
Comparative plant genomics examines the similarities of, and differences in, genomes between plant species. By comparing genomes of evolutionarily divergent species, we can better understand the patterns and processes that underlie plant genome evolution as well as uncover functional regions of genomes [7]. Structural variations are large (> 1 kbp in size) rearrangements of DNA that include insertions, deletions, duplications (also referred to as copy number variations), inversions, and translocations [8]. These genomic alterations are an important source of genetic and phenotypic diversity. For example, structural variations in plants have been associated with stress tolerance, disease resistance, domestication, and increase in yields, leaf size, fruit shape, reproductive morphology, adaptation, and speciation [9]. Through the use of cytogenetics, researchers have been able to identify large chromosomal changes (e.g., translocations, aneuploidy, and loss of repeats) [10], yet this method is labor intensive, prone to error, and generally only captures differences with limited resolution. Thus, cytogenetic methods may greatly underestimate the number of diverse changes in architecture that are in fact found between plant genomes.
As sequencing technologies have become more accessible, the field of comparative genomics has greatly expanded our knowledge of plant genome structure. With the high throughput and low cost of next-generation sequencing (NGS), more than 100 plant genomes have been sequenced. Although short-read sequencing is useful for detecting small-scale sequence variations (e.g., < 1 kbp in size or a few nucleotides), it is unable to detect most large-scale structural variations. Plant genomes are notorious for being large and highly repetitive, and many contain multiple copies of entire chromosomes (e.g., polyploidy).
Most sequencing techniques are effective at detecting deletions but have difficulty resolving sequence redundancies owing to short reads typical of NGS. Thus, sequencing advances alone may not provide sufficient resolution for comparing the organization and structure of genomes from evolutionarily divergent species [2] (Figure 1).
Structural variations are large (> 1 kbp) rearrangements of DNA that frequently result in phenotypic differences. These variations include insertions, deletions, inversions, duplications, and translocations. By comparing genomes of different species, large chromosomal changes can be identified.
Mapping genomes
Physical maps: One method to compare genome structures is physical maps [2]. Just as a cartographer would start with key landmarks when mapping a region, then later fill in the details of the location, physical maps provide molecular anchor points to link sequence contigs, bridge repetitive regions and give a course-grain view of genome structure. Physical maps have been key in completing high-quality genome assemblies and are typically made using large insert clone libraries, such as bacterial artificial chromosomes (BACs). Although BAC-based physical maps are helpful in the completion of de novo genome sequencing, their widespread use in plant comparative genomics has been limited because they are expensive and time-consuming, and require a great deal of experimental expertise. Additionally, BAC libraries are subject to clone amplification biases resulting in incomplete coverage, and some regions of BAC physical maps can be difficult to resolve due to the sequence redundancy typically found in plant genomes [11].
Tical maps: An optical map is an ordered genome-wide physical map constructed from unamplified DNA molecules. A unique ‘fingerprint’ or ‘barcode’ is created by mapping the location of restriction enzyme recognition sites present in a long DNA molecule. Optical mapping has advantages over other genomic technologies because it uses long DNA molecules that are not cloned or amplified and preserves the order of restriction enzyme recognition sites. This allows a map to be created that accurately reflects long repetitive regions, and is free from cloning or amplification bias. It also allows the map to resolve complicated genome regions, including copy number variations and potentially homoeologous segments from polyploid genomes, more efficiently and unambiguously than unordered restriction fragment maps. Although this system has been useful in improving plant genome assemblies [10], it was initially applied to small genomes (e.g. fungi and bacteria). The low throughput of traditional optical mapping makes it difficult to use in large-scale plant comparative genomics projects.
Genome mapping: Through advances in labeling, imaging, automation, and nanofabrication, a higher-throughput mapping system has recently been developed. This mapping system has been commercialized by BioNano Genomics as the Irys platform. Its capabilities to capture 50–200 gbp of data per day have led to its increasing popularity among researchers. The relative ease of quickly mapping large genomes at high coverage to identify structural variations with or without a reference sequence assembly suggests that this system has potential for use in plant comparative genomics.
Data Collection in comparative genome mapping
The first step to creating a genome map is to collect high-quality data. Data quality is partly determined by individual molecule length and the accuracy of the measured distances between nick-repaired fluorescent labels. The use of high-molecular-weight (HMW) DNA allows a single DNA molecule to cover long genomic regions, often spanning problematic regions of the genome that have been difficult to resolve using NGS. However, the physical isolation of long, high-quality DNA in plants can be challenging due to the variety of organic compounds plant tissues contain. Plants harbor large amounts of polyphenols, polysaccharides, and other secondary metabolites, which normally aid in functions such as plant defenses, but can also contaminate DNA for laboratory purposes [12]. Different species, different parts of a plant and even the same plant at different development stages can all vary in their chemical composition, making protocols problematic to generalize. One way to avoid excess secondary metabolites is to use very young tissue; new tissue has the highest DNA content to mass ratio possible, and the lowest level of metabolites. Dark treating seedlings for 24-72h significantly reduces the number of polysaccharides and has shown favorable results [13]. Although only a small amount of DNA is needed (as little as 300 ng), isolating HMW DNA from plants can be challenging and methods are not as well established as conventional DNA extractions. The isolation of intact nuclei can keep the DNA from shearing after the disruption of the cell wall. Isolated nuclei can also be cleaned extensively to remove contaminants that are generally in the cytoplasm [14]. Contaminants are removed through the use of a strong reducing agent (e.g., 2-mercaptoethanol), detergents (e.g., Triton-X 100), and polymers e.g., polyvinyl-pyrrolidone (PVP) that prevent the oxidation of polyphenols, solubilize lipids and enzymes and bind polyphenols, all of which can reduce DNA quality.
Furthermore, a Percoll density gradient is used to help separate nuclei from particles of a different density. After isolation, nuclei are washed several times before being embedded in low-melting-point agarose plugs before membrane lysis. The physical matrix of agarose plugs allows the naked DNA to be further treated with RNase and proteinases, and to be washed to remove contaminants and residual reagents without excessive physical shearing of the DNA. Extraction of DNA from the plugs requires melting the agarose plug and subsequent drop dialysis, which further removes low-molecular-weight contaminants and salts, as well as concentrating the DNA [15]. Once enough sufficiently pure DNA has been isolated, the actual mapping can begin.
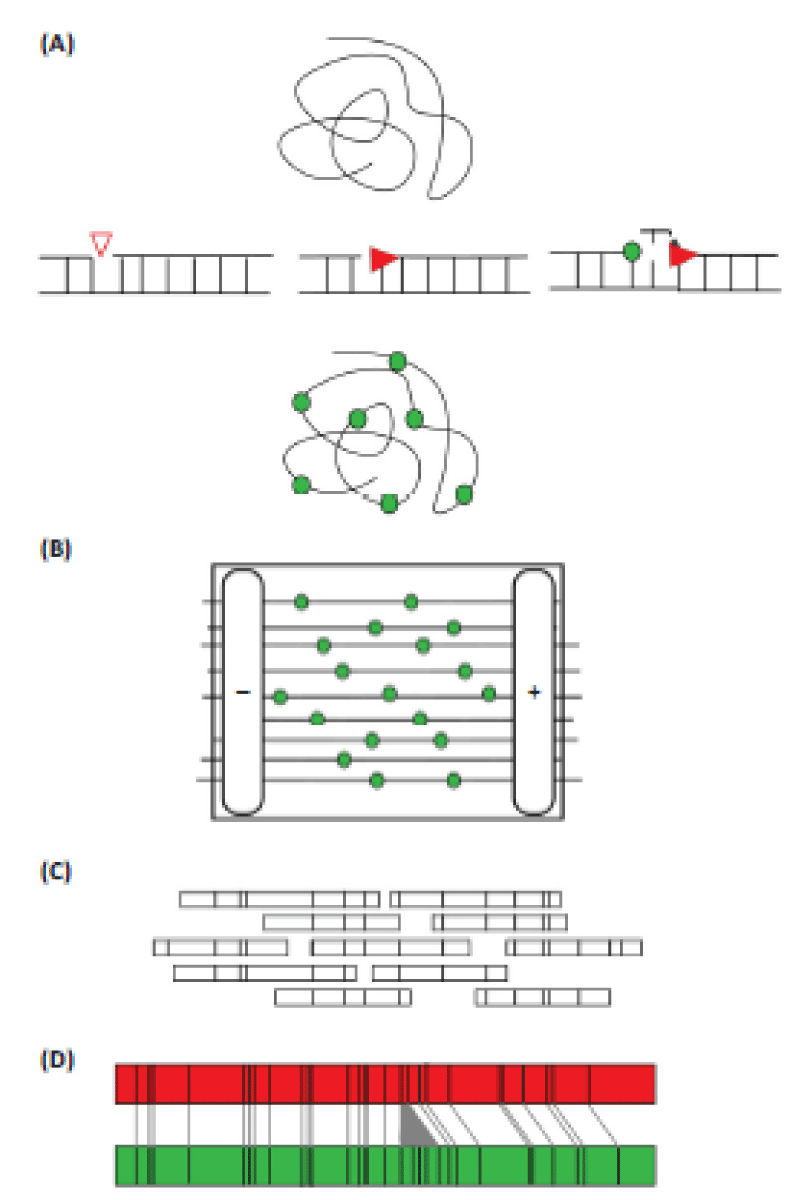
Sequence-specific, single-strand breaks or nicks are introduced into the DNA with a modified restriction enzyme or nickase. A polymerase then incorporates fluorescent nucleotide analogs at the break sites. Labeled DNA is loaded onto a nanofluidic chip, where an automatically applied electric field draws iterative samples of DNA through a series of columns for linearization, and then into nanochannels for imaging. Electric currents are applied in such a way that the large pool of HMW DNA is repeatedly sampled through a different series of run cycles. Once labeled DNA from the first cycle is positioned in the channel, fragments are imaged through an automated system on the Irys instrument [3]. A series of raw images are converted to single molecule maps, digital measurements of molecule length and intensity, as well as physical distances between and intensity of incorporated labels (Figure 2).
A) Data collection: high-molecular-weight (HMW) DNA is extracted and a single-stranded break or nick is introduced at a sequence-specific recognition site on individual DNA molecules. DNA is fluorescently labeled at the sequence-specific site using a DNA polymerase enzyme and nucleotide. The DNA backbone is stained to allow for accurate measurement of the DNA molecule.
B) On a specialized chip, automated electrophoresis pulls DNA into arrays of nanochannels where linearized DNA is imaged.
C) Data analysis: each imaged molecule is digitally measured for length and distances between labeled sites to create a molecule map. Molecule maps that overlap and whose distance patterns match are assembled into a consensus contig.
D) Consensus contig maps from different plant samples are then compared to identify large structural variations, such as an insertion in the green contig depicted here.
Data analysis
Creating a high-quality consensus map: Once data have been collected, single-molecule maps are assembled into a consensus map that spans a large genomic region. Each imaged molecule is characterized by its total length estimate and a linear series of fluorescently labeled nick sites that represent physical distances between endonuclease recognition sites and each ‘fragment pattern’ matches a distinct region of the genome. These distinct series of kilobase-sized distances are analogous to fragments from digested BACs, except that they are already arranged in linear order. Molecule maps that overlap is identified through a heuristic alignment algorithm that first matches partial distance patterns. Consensus contigs are created using an overlap-layout-consensus algorithm. The end result of the assembly process is a set of contigs with unique distance patterns, each of which represents a certain region of a chromosome within the plant genome, and is referred to as a consensus genome map.
Typical of other genomic approaches, contigs representing intact entire chromosomes are not usually achieved; however, a low number of long contigs that match the expected genome size and chromosome number would suggest complete assembly. There are several factors that may impact assembly quality, including DNA quality, nick efficiency, imaging artifacts, and genome complexity. Common errors in these data include: (i) inaccurate sizing of molecules due to non-uniform fluorescent staining or stretching; (ii) spurious enzyme cut sites due to random breakage of the DNA molecule or star activity (false-positive label sites); and (iii) missing label sites due to missing enzyme cut sites, incomplete digestion, or labeling errors (false-negative label sites). Traditional optical mapping, OpGen, also has the added error of small fragment loss [16]. While careful laboratory techniques aim to minimize the impact of these factors, they cannot be entirely removed. Thus, part of the assembly quality depends on the effectiveness of the assembly algorithm at compensating for noise in the input data.
Algorithms and resources
To align single-molecule maps into a consensus genome map, dynamic programming algorithms are used to account for the inherent error characteristics of the molecule maps. Many of the methods and approaches used by common DNA sequence assembly programs are not useful for imaged HMW molecules due to differences in how data are generated (e.g., not amplified, single ‘dimension’ of fragment length) [17]. Compared with the analytical advances made in sequencing data, there are relatively few methods that exist for analyzing and utilizing genome map data. There have been a series of software tools developed specifically for the BioNano Irys system to improve mapping quality. The Irys Solve software addresses noise in data by allowing users to customize many input parameters that describe the error profile for their data, such as false positive and false negative nicks, molecule stretch, and fluorescence intensity, which can be estimated empirically using a genome sequence assembly. The algorithm then makes compensatory decisions based on those input parameters. Furthermore, Irys Solve has been developed to detect a variety of structural variants in large genomes (e.g., humans). However, to date, there has only been experimental validation of its ability to detect insertions and deletions; other structural variants, such as inversions or translocations, have yet to be validated.
Sharp et al. developed a method that selects the best input parameters by running multiple assemblies with permutations of input parameters with the minimal resource (computing) using tools developed by Shelton et al. primarily focus on complementing genome sequence assembly. One tool maps a subset of the molecules to an in silico digest of a reference sequence assembly at a variety of error profiles, selecting the profile that maximizes the mapping efficiency [18]. Additionally, they present software called Stitch that automatically parses and interprets the output from a comparison between a consensus map and a reference genome to super scaffold a sequence assembly [18]. Another piece of software, ALL MAPS, performs a similar computational task as Stitch to link map data with draft genome sequence assemblies, although it is not written specifically for genome map data [19]. Most other software is not written specifically for the Irys genome-mapping system but can be used on both traditional optical mapping and Irys genome mapping data. Some existing algorithms, which were originally developed for small genomes, are unable to be ported to large genome assemblies because of computational limitations.
For example, Genting, a proprietary software, has been able to successfully assemble small consensus optical maps but is unable to scale to large genomes. Opgen’s MapSolver software allows for map visualization, comparisons between maps or between a map and an in silico digest of a sequence assembly, and aids in assembly improvement, but only on genomes up to 100 mbp, ruling out most plants. Its Genome-Builder software uses an iterative Bayesian maximum-likelihood, a modified Smith-Waterman dynamic algorithm, plus a heuristic filtering process. This software is capable of performing assembly improvement (super-scaffolding) but does not generate a consensus map; neither does it facilitate direct comparisons between optical maps from two plants [16]. There have been several programs that have the computational feasibility for large genome map assemblies. For example, Valouev et al. developed an algorithm that is able to align two optical maps and also align an optical map to a reference map. This is the first algorithm capable of producing accurate maps of large genomes in a feasible timeframe. SOMA and TWIN are both open-source software that aligns optical map data to a reference sequence, but the latter is highly sensitive to false positive and false negative label sites in the input data [20].
Utilizing optical map assemblies
Once high-quality consensus maps have been created, several downstream analyses may be performed. Current applications of genome mapping have primarily been used to improve or validate sequence assemblies (e.g., to improve the resolution of contigs from BAC pools of wheat 7D short arm and one of the most contiguous de novo assemblies of a human genome. However, genome mapping could be used to replace several well-established but low-throughput technologies for comparative genomics; furthermore, it is superior to sequencing to detect large structural variants because of large input molecules. The use of genome-mapping comparisons has not been fully utilized in plant genomes due to historically high costs [19].
Structural variations between species
Genome mapping has the potential to be used for comparing structural variations between species. Structural variations are thought to be the major contributors to phenotypic variation in plants, leading to an increased focus on characterizing structural variations between plant genomes. One example of the potential use of optical mapping to compare structural variation across plant species is in Brassica napus.
Segregating populations of B. napus doubled haploid lines and codominant RFLP markers detected pairs of homoeologous loci on N7 and N16 for which the annual and biennial parents had identical alleles in regions expected to be homoeologous. High-throughput genome mapping could replace labor-intensive RFLP or in situ hybridization experiments to more quickly uncover such large genomic rearrangements. Analysis of intentionally derived genetic stocks, for example, the wheat nullisomic-tetrasomic lines or chromosome arm deletion stocks, have been used to physically locate the position of genes on chromosome arms [21]. Genome mapping could be used on these same genetic stocks to further identify breakpoints and missing genomic regions.
Understanding the evolution of genomes in polyploidy
Polyploidy, the doubling of all the chromosomes in a cell, is ubiquitous in the evolution of plant species. Most, if not all, angiosperm species have gone through multiple rounds of polyploid- ization. At the onset of polyploidization, a period of rapid genomic reorganization and massive gene loss occurs and structural variations arise [22]. Structural variations can also arise through local duplication events and the activity of transposons, resulting in the differential loss of genes between lineages. Little is known about how long chromosomal variation may persist and how it might influence the establishment and evolution of polyploids in nature. Genome mapping could be used to characterize chromosomal composition before and after polyploidization events. For example, cultivated cotton is an allotetraploid that evolved following a polyploidization event involving two diploid kinds of cotton (the A- and D-genomes) 1–2 million years ago. The cotton system serves as an excellent model for identifying structural variations between species, more specifically, examining nonreciprocal homologous recombination, the intergenomic spread of transposable elements, and alterations and biases in gene duplicate expression. Research has shown some evidence of chromosomal rearrangements between the homoeologous genomes in polyploids [22], but it is still not completely understood how genome variations compare across different cotton species and genomes. Genome mapping could be useful in pinpointing segmental losses and exchanges among homoeologous chromosomes, which are important aspects of polyploidy genome evolution [19].
Use of comparative genome mapping in crop improvement
One common use of comparative genomics is in plant breeding for crop improvement. Alleles that differ between lines can be correlated with favorable agronomic traits. While it is theoretically feasible to incorporate genome mapping for structural variant genotyping into this system, there are some practical problems that make the idea less tenable. The coverage required for structural-variant calling using map fingerprints is likely lower than that required for whole-genome assembly, but it is unclear what the required coverage would be. Also, the number of different lines that are normally required for a large-scale breeding project would currently be prohibitive, because genome-mapping technology has not yet been optimized to run several samples concurrently. However, the future of crop improvement will likely be centered on a deep functional understanding of the genome of a species, including structural variants [23], in which case genome mapping could help address key biological questions for crop improvement.
Future perspectives of comparative genome mapping in plant breeding
Future developments in genome mapping include multicolored labeling that will allow the recognition of multiple sequence motifs in a single sample [24]. The ability to map the epigenome through labeling DNA methylation will allow the comparison of the genetic and epigenetic composition of genomes [25]. If genome mapping is to complement DNA sequencing, parallelizing data collection by both technologies is required. Past plant comparative genomic studies have investigated differences in genome size, gene number, transposable elements, and syntenic relations, yet their methods underestimate the diverse architecture found in plant genomes [7].
Many studies that address changes in genomic content focus on single-nucleotide polymorphisms or short indels as markers of association genetics, yet this research largely ignores the large structural variations that often have significant impacts. Direct comparisons of large genomic structural variations have so far been lacking in plants, and genome mapping shows great promise for revealing genomic regions that are not easily accessible through conventional sequencing methods. Genome mapping could become an integral tool in the study of plant domestication, polyploid evolution, and trait development [25-27].
Conclusion
Through the use of genome mapping, large gains in the field of plant comparative genomics are likely. The long-range information that is able to span complex genomic regions will greatly improve our understanding of relations between different genomes. Furthermore, the ability to capture large quantities of data in a relatively short time frame and at a low cost will allow researchers to compare whole genomes of multiple plant species with relative ease. These qualities of genome mapping make it an extremely useful tool in situations where a low-resolution genomic picture is sufficient, such as identifying structural variations between plant species and identifying phylogenetic patterns in genome evolution. However, for this technology to reach its full potential, obstacles must be overcome.
- Paterson AH, Bowers JE, Burow MD, Draye X, Elsik CG, Jiang CX, Katsar CS, Lan TH, Lin YR, Ming R, Wright RJ. Comparative genomics of plant chromosomes. Plant Cell. 2000 Sep;12(9):1523-40. doi: 10.1105/tpc.12.9.1523. PMID: 11006329; PMCID: PMC149067.
- Lewin HA, Larkin DM, Pontius J, O'Brien SJ. Every genome sequence needs a good map. Genome Res. 2009 Nov;19(11):1925-8. doi: 10.1101/gr.094557.109. Epub 2009 Jul 13. PMID: 19596977; PMCID: PMC2775595.
- Lam ET, Hastie A, Lin C, Ehrlich D, Das SK, Austin MD, Deshpande P, Cao H, Nagarajan N, Xiao M, Kwok PY. Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat Biotechnol. 2012 Aug;30(8):771-6. doi: 10.1038/nbt.2303. PMID: 22797562; PMCID: PMC3817024.
- Vavilov NI. The law of homologous series in variation. J Genet. 1922; 12: 1.
- Tanksley SD, Bernatzky R, Lapitan NL, Prince JP. Conservation of gene repertoire but not gene order in pepper and tomato. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6419-23. doi: 10.1073/pnas.85.17.6419. PMID: 16593975; PMCID: PMC281983.
- Allen P. Everything Old is New Again. A&M Recods. 1974.
- Caicedo AL, Purugganan MD. Comparative plant genomics. Frontiers and prospects. Plant Physiol. 2005 Jun;138(2):545-7. doi: 10.1104/pp.104.900148. PMID: 15955910; PMCID: PMC1150366.
- Gaut BS, Wright SI, Rizzon C, Dvorak J, Anderson LK. Recombination: an underappreciated factor in the evolution of plant genomes. Nat Rev Genet. 2007 Jan;8(1):77-84. doi: 10.1038/nrg1970. PMID: 17173059.
- Lowry DB, Willis JH. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol. 2010 Sep 28;8(9):e1000500. doi: 10.1371/journal.pbio.1000500. Erratum in: PLoS Biol. 2012 Jan;10(1):10.1371/annotation/caa1b7dd-9b6d-44db-b6ce-666954903625. PMID: 20927411; PMCID: PMC2946948.
- Tang H, Krishnakumar V, Bidwell S, Rosen B, Chan A, Zhou S, Gentzbittel L, Childs KL, Yandell M, Gundlach H, Mayer KF, Schwartz DC, Town CD. An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genomics. 2014 Apr 27;15:312. doi: 10.1186/1471-2164-15-312. PMID: 24767513; PMCID: PMC4234490.
- Meyers BC, Scalabrin S, Morgante M. Mapping and sequencing complex genomes: let's get physical! Nat Rev Genet. 2004 Aug;5(8):578-88. doi: 10.1038/nrg1404. PMID: 15266340.
- Sahu SK, Thangaraj M, Kathiresan K. DNA Extraction Protocol for Plants with High Levels of Secondary Metabolites and Polysaccharides without Using Liquid Nitrogen and Phenol. ISRN Mol Biol. 2012 Nov 14;2012:205049. doi: 10.5402/2012/205049. PMID: 27335662; PMCID: PMC4890884.
- Hein I, Williamson S, Russell J, Powell W. Isolation of high molecular weight DNA suitable for BAC library construction from woody perennial soft-fruit species. Biotechniques. 2005 Jan;38(1):69-71. doi: 10.2144/05381ST02. PMID: 15679088.
- Zhang M, Zhang Y, Scheuring CF, Wu CC, Dong JJ, Zhang HB. Preparation of megabase-sized DNA from a variety of organisms using the nuclei method for advanced genomics research. Nat Protoc. 2012 Feb 16;7(3):467-78. doi: 10.1038/nprot.2011.455. PMID: 22343429.
- Marusyk R, Sergeant A. A simple method for dialysis of small-volume samples. Anal Biochem. 1980 Jul 1;105(2):403-4. doi: 10.1016/0003-2697(80)90477-7. PMID: 7457844.
- Dong Y, Xie M, Jiang Y, Xiao N, Du X, Zhang W, Tosser-Klopp G, Wang J, Yang S, Liang J, Chen W, Chen J, Zeng P, Hou Y, Bian C, Pan S, Li Y, Liu X, Wang W, Servin B, Sayre B, Zhu B, Sweeney D, Moore R, Nie W, Shen Y, Zhao R, Zhang G, Li J, Faraut T, Womack J, Zhang Y, Kijas J, Cockett N, Xu X, Zhao S, Wang J, Wang W. Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus). Nat Biotechnol. 2013 Feb;31(2):135-41. doi: 10.1038/nbt.2478. Epub 2012 Dec 23. PMID: 23263233.
- Valouev A, Li L, Liu YC, Schwartz DC, Yang Y, Zhang Y, Waterman MS. Alignment of optical maps. J Comput Biol. 2006 Mar;13(2):442-62. doi: 10.1089/cmb.2006.13.442. PMID: 16597251.
- Shelton JM, Coleman MC, Herndon N, Lu N, Lam ET, Anantharaman T, Sheth P, Brown SJ. Tools and pipelines for BioNano data: molecule assembly pipeline and FASTA super scaffolding tool. BMC Genomics. 2015 Sep 29;16:734. doi: 10.1186/s12864-015-1911-8. PMID: 26416786; PMCID: PMC4587741.
- Tang H, Zhang X, Miao C, Zhang J, Ming R, Schnable JC, Schnable PS, Lyons E, Lu J. ALLMAPS: robust scaffold ordering based on multiple maps. Genome Biol. 2015 Jan 13;16(1):3. doi: 10.1186/s13059-014-0573-1. PMID: 25583564; PMCID: PMC4305236.
- Lin HC, Goldstein S, Mendelowitz L, Zhou S, Wetzel J, Schwartz DC, Pop M. AGORA: Assembly Guided by Optical Restriction Alignment. BMC Bioinformatics. 2012 Aug 2;13:189. doi: 10.1186/1471-2105-13-189. PMID: 22856673; PMCID: PMC3431216.
- Law CN. Aneuploidy in wheat and its uses in genetic analysis. In Wheat Breeding (Lupton, D.F.G.H., ed.). 1987; 71–108.
- Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, Zhang J, Saski CA, Scheffler BE, Stelly DM, Hulse-Kemp AM, Wan Q, Liu B, Liu C, Wang S, Pan M, Wang Y, Wang D, Ye W, Chang L, Zhang W, Song Q, Kirkbride RC, Chen X, Dennis E, Llewellyn DJ, Peterson DG, Thaxton P, Jones DC, Wang Q, Xu X, Zhang H, Wu H, Zhou L, Mei G, Chen S, Tian Y, Xiang D, Li X, Ding J, Zuo Q, Tao L, Liu Y, Li J, Lin Y, Hui Y, Cao Z, Cai C, Zhu X, Jiang Z, Zhou B, Guo W, Li R, Chen ZJ. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol. 2015 May;33(5):531-7. doi: 10.1038/nbt.3207. Epub 2015 Apr 20. PMID: 25893781.
- Morrell PL, Buckler ES, Ross-Ibarra J. Crop genomics: advances and applications. Nat Rev Genet. 2011 Dec 29;13(2):85-96. doi: 10.1038/nrg3097. PMID: 22207165.
- Hastie AR, Dong L, Smith A, Finklestein J, Lam ET, Huo N, Cao H, Kwok PY, Deal KR, Dvorak J, Luo MC, Gu Y, Xiao M. Rapid genome mapping in nanochannel arrays for highly complete and accurate de novo sequence assembly of the complex Aegilops tauschii genome. PLoS One. 2013;8(2):e55864. doi: 10.1371/journal.pone.0055864. Epub 2013 Feb 6. PMID: 23405223; PMCID: PMC3566107.
- Levy-Sakin M, Grunwald A, Kim S, Gassman NR, Gottfried A, Antelman J, Kim Y, Ho SO, Samuel R, Michalet X, Lin RR, Dertinger T, Kim AS, Chung S, Colyer RA, Weinhold E, Weiss S, Ebenstein Y. Toward single-molecule optical mapping of the epigenome. ACS Nano. 2014 Jan 28;8(1):14-26. doi: 10.1021/nn4050694. Epub 2013 Dec 20. PMID: 24328256; PMCID: PMC4022788.
- Das SK, Austin MD, Akana MC, Deshpande P, Cao H, Xiao M. Single molecule linear analysis of DNA in nano-channel labeled with sequence specific fluorescent probes. Nucleic Acids Res. 2010 Oct;38(18):e177. doi: 10.1093/nar/gkq673. Epub 2010 Aug 10. PMID: 20699272; PMCID: PMC2952877.
- Tang Z, Li M, Chen L, Wang Y, Ren Z, Fu S. New types of wheat chromosomal structural variations in derivatives of wheat-rye hybrids. PLoS One. 2014 Oct 10;9(10):e110282. doi: 10.1371/journal.pone.0110282. PMID: 25302962; PMCID: PMC4193885.
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
