International Journal of Agricultural Science and Food Technology
AMMI with BLUP analysis for stability assessment of wheat genotypes under multi locations timely sown trials in Central Zone of India
Ajay Verma* and GP Singh
Cite this as
Verma A, Singh GP (2021) AMMI with BLUP analysis for stability assessment of wheat genotypes under multi locations timely sown trials in Central Zone of India. J Agric Sc Food Technol 7(1): 118-124 DOI: 10.17352/2455-815X.000098AMMI analysis explained the highly significant effects of the environment, GxE interaction, and genotypes for wheat genotypes evaluated under restricted irriation timely sown multi-location trials in the Central zone of the country during 2018-19 and 2019-20. About 77.1%, 12.2% & 2.3% had been contributed by environments, interactions and genotypes of the total sum of squares due to treatments respectively in the first year. The utilization of more number of IPCA’s in AMMI and WAASB stability measures had altered the ranking of genotypes. Analytic measures of adaptability and Superiority indexes as per BLUP of genotypes identified DBW110, MP3288. Adaptability measures as per arithmetic, geometric and harmonic means and their corresponding values expressed deviation as observed in a seperate quadrant of Biplot graphical analysis. However, this group maintained the right angle with MASV, MASV1, and stability measures. The cluster of Superiority indexes as per averages yield of wheat genotypes placed in the adjacent quadrant. Superiority indexes favored HI8823, MP3288, DBW110 wheat genotypes for high yield and stable performance for the second year. Adaptability measures as per arithmetic, geometric and harmonic means along with the corresponding values of RPGV & MHRPGV expressed bondage and placed in a different quadrant. Cluster of Superiority indexes as per averages of the yield of wheat genotypes seen in the same quadrant as more than 73.5% variation accounted for by the first two principal components.
Introduction
AMMI analysis had been employed mostly to have an efficient estimation of GxE interactions crop breeding trials even large number of other statistical procedures for the stability analyses has been validated in literature [1]. Recently the effects of genotypes, environments, or both to be advocated as of random nature [2]. BLUP have proved its potential to improve the predictive accuracy of random effects under mixed model approach [3]. Both BLUP and AMMI, approaches, seperated the pattern from the random error components in GxE interactions analysis [4]. Integration of stability of performance with yield, is necessary for selecting high yielding, stable genotypes [5]. Both yield and stability of performance should be considered simulaneously, to reduce the effect of G x E interaction and make selection more precise and reliable [6]. These two approaches have been used separately in the field evaluation of genotypes under multi location trials [7]. Benefits of these two important techniques, AMMI and BLUP, utilized to define Superiority Index measure for the stability and adaptability of genotypes [5]. The current study dealt with the analysis of G x E interaction and yield stability through AMMI with BLUP techniques for evaluated wheat genotypes.
Materials and methods
States of India comprised by Madhya Pradesh, Chhattisgarh, Gujarat, Rajasthan (Kota and Udaipur divisions) and Jhansi division of Uttar Pradesh is known as Central Zone. This zone is well established for the quality products of wheat especially chapatti in the country and abroad. Six promising wheat genotypes in advanced trials evaluated at twelve major locations of the zone and eight genotypes at thirteen locations during 2018-19 and 2019-20 cropping seasons respectively. Field trials were conducted at research centers in randomized complete block designs with three replications. Recommended agronomic practices were followed to harvest good yield. Details of genotype parentage along with environmental conditions were reflected in Tables 1,2 for ready reference.
Stability measure as Weighted Average of Absolute Scores has been calculated as
Where WAASBi was the weighted average of absolute scores of the ith genotype (or environment); IPCAik the score of the ith genotype (or environment) in the kth IPCA, and EPk was the amount of the variance explained by the kth IPCA. Superiority index allowed variable weights to yield and stability measure (WAASB) to select genotypes that combined high performance and stability as where rGi and rWi were the rescaled values for yield and WAASB, respectively, for the ith genotype; Gi and Wi were the yield and the WAASB values for ith genotype. SI Superiority index for the ith genotype that weighted between yield and stability, θY and θS were the weights for yield and stability assumed to be of order 65 and 35 respectively in this study,
AMMI analysis was performed using AMMISOFT version 1.0, available at https://scs.cals.cornell.edu/people/ hugh-gauch/ and SAS software version 9.3. Stability measures had been compared with recent analytic measures of adaptability calculated as the relative performance of genetic values (PRVG) and harmonic mean based measure of the relative performance of the genotypic values (MHPRVG) for the simultaneous analysis of stability, adaptability, and yield [11].
Results and discussion
First-year 2018-19
Environment (E), Genotypes (G), and GxE interaction effects were highly significant as mentioned by the AMMI analysis (Table 3). Analysis observed the greater contribution of environments, GxE interactions, and genotypes to the total sum of squares (SS) as compared to the residual effects. Further SS attributable to GxE interactions was partitioned as attributed to GxE interactions Signal and GxE interactions Noise. AMMI analysis was appropriate for data sets where-in SS due to were of magnitude at least of due to additive genotype main effects [12]. The SS for GxE interactions Signal was higher compared to genotype main effects, indicated appropriateness of AMMI analysis. The environment significantly explained about 77.1% of the total sum of squares due to treatments indicating that diverse environments caused most of the variations in genotypes yield [13]. Genotypes explained only 2.3% of the total sum of squares, whereas GxE interaction accounted for 12.2% of treatment variations in yield. First four significant multiplicative terms explained 94.7 % of interactions sum of squares and the remaining 5.3% was the discarded residual [14].
Ranking of genotypes vis-à-vis number of IPCA’s
The IPCA scores of genotypes in the AMMI analysis were an indication of stability or adaptability over environments. The greater the IPCA scores, the more specific adapteded genotype to certain locations. The more the IPCA scores approximate to zero, the more stable or adapted the genotypes is overall the locations. The ranking of genotype as per absolute IPCA-1 scores were DDW47, MP3288 (Table 4). While for IPCA-2, genotypes DBW277, DDW47 would be of choice. Values of IPCA-3 favored DBW277, DBW110 wheat genotypes. As per IPCA-4, DDW47, DBW110 genotypes would be of stable performance. Analytic measures of adaptability MASV and MASV1 consider all significant IPCAs of the analysis. Genotypes DDW47, DBW277 had been identified by MASV & MASV1 measures (Ajay et al., 2019). To identify how the ranks of evaluated wheat genotype altered with utilizing numbers of IPCA in the WAASB estimation, the genotype’s ranks were obtained while considering 1, 2,..., p IPCA’s in the WAASB calculations. WAASB = |IPCA1| for using only first IPCA. The genotype with the smallest WAASB value had been ranked with the first-order. Preferences of wheat genotypes as per W1, W2 & W3 measures were the same as DDW47, MP3288 identified as two promising genotypes through the higher-order varied from these measures. Stability measure WAASB based on all significant IPCA’s settled for DDW47, MP3288 genotypes for considered locations of the zone for stable high yield. The genotypes ranking was altered by the extent to which IPCAs were included in the WAASB estimation. This reinforces the benefits of using the WAASB index since it captures the variations of all IPCAs to compute the stability [5].
Stable and productive génotypes by AMMI & BLUP
An average yield of genotypes as per BLUP values of genotypes yield selected DBW110, DBW277 wheat genotypes (Table 5). This method is simple, but not fully exploiting all information contained in the dataset. A geometric mean is used to evaluate the adaptability of genotypes and genotypes with high GM will be desirable. Geometric mean top-ranked DBW110, MP3288 genotypes. As proposed by Resende [15], a method to rank genotypes considering the yield and stability simultaneously is the harmonic mean of genetic values (HMGV). In the context of mixed models, the Harmonic Mean of Genotypic Values were calculated as genotypes with greater values would be recommended. Harmonic Mean expressed higher values for DBW110, MP3288 genotypes. Moreover, the Harmonic Mean of Relative Performance of Genotypic Values (HMRPGV) method proposed by Resende [15] that used Restricted Maximum Likelihood (REML) or Best Linear Unbiased Prediction (BLUP) as similar to the methods of Lin and Binns [16] and Annicchiarico [17]. In the HMRPGV method for stability analysis, the genotypes can be simultaneously sorted by genotypic values (yield) and stability using the harmonic means of the yield so that the smaller the standard deviation of genotypic performance among the locations. Values of HMRPGV ranked DBW110, MP3288 the performance of the genotypes among the locations. When considering the yield and adaptability simultaneously, the recommended approach is the relative performance of genetic values (RPGV) overcrop years. For adaptability analysis, the Relative Performance of Genotypic Values had been measured across environments and settled for DBW110, MP3288 wheat genotypes.
While assigning 65 and 35 weights to average yield (AM) and stability, the Superiority index pointed out DBW110, MP3288 genotypes would maintain high yield and stable performance. SI measure, considered GM and stability, selected DBW110, MP3288 genotypes. Values of SI, using HM and stability, favored the same set of wheat genotypes DBW110, MP3288. Analytic measures of adaptability RPGV and MHRPGV pointed out DBW110, MP3288 would be more adaptable genotypes.
Biplot analysis of measures
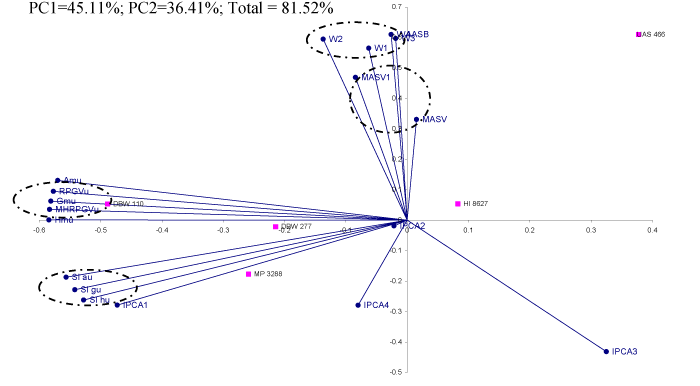
Table 6 reflected approximately 81.5% of the total variation with 45.1% & 36.4 % contributions of the first two significant PC’s [18]. A group comprised of MASV, MASV1 & stability measures by utilizing the number of interaction principal components (Figure 1). Adaptability measures as per arithmetic, geometric and harmonic means along with the corresponding values of RPGV & MHRPGV expressed bondage and placed together in a different quadrant. However, this group maintained right angle with MASV, MASV1 & stability measures. The cluster of Superiority indexes as per averages of the yield of wheat genotypes seen in a different quadrant. Moreover, the performance of genotypes would be more or less the same by Superiority indexes and adaptability measures as acute angles observed in corresponding rays.
Second-year 2019-20
Environment (E), genotypes (G) and GxE interaction effects were highly significant as mentioned by the AMMI analysis (Table 3). The environment significantly explained about 70.1%, GxE interaction accounted for 14.2% and genotypes accounted for only 1.3% of the total sum of squares. Significant six multiplicative terms explained 98.7 % and the remaining 1.3% residual was discarded.
Ranking of genotypes vis-à-vis number of IPCA’s
The preference order of genotypes as per IPCA-1 scores was DDW47, UAS466, HI8823 (Table 7). While the values of IPCA-2 selected MP3288, UAS466, DBW110 genotypes would be of choice. Values of IPCA-3 favored DBW110, MP3288, UAS472 wheat genotypes. As per IPCA-4, UAS472, DDW47, HI8627 genotypes would be of stable performance. DBW110, HI8627, MPO1357, genotypes pointed by IPCA-5 measure. Genotypes HI8823, MPO1357, DDW47 were identified by absolute values of IPCA-6. Analytic MASV and MASV1 measures of adaptability considered all significant IPCAs of the analysis simultaneously. MASV1 identified genotypes UAS466, HI8627, MP3288 would express stable yield whereas genotypes HI8627, UAS466, UAS472 be of stable performance by MASV measure respectively.
Genotype preferences varied from DDW47, UAS466, HI8823 based on W1 whereas UAS466, DDW47, HI8627 as per W2 values while UAS466, DDW47, HI8627 by values of W3 (Table 8). Genotypes UAS466, DDW47, HI8627 were pointed by W4; W5 favored UAS466, DDW47, HI8627. Stability measure WAASB based on all significant IPCA’s settled for UAS466, DDW47, HI8627 genotypes for considered locations of the zone for stable high yield. The genotype ranking was altered by the extent to the number of IPCAs included in the WAASB estimation. This reinforced the benefits of using the WAASB index since it captures the variations of all IPCAs to compute the stability.
Stable and Productive genotypes by AMMI & BLUP
Average yield based on BLUP values of genotypes selected HI8823, MP3288, DBW110 wheat genotypes (Table 9). Geometric mean observed HI8823, MP3288, DBW110 were top-ranked genotypes. Harmonic Mean of yield expressed higher values for MP3288, HI8823, DBW110 genotypes. Values of HMRPGV ranked MP3288, HI8823, DBW110 the performance of the genotypes among the locations. Relative Performance of Genotypic Values had settled for HI8823, MP3288, DBW110 wheat genotypes.
While assigning 65 and 35 weights to average yield (AM) and stability, the Superiority index pointed out HI8823, MP3288, DBW110 genotypes would maintain high yield and stable performance. SI measure considered GM and stability, selected HI8823, MP3288, DBW110 genotypes. Values of SI, using HM and stability, favored the same set of wheat genotypes HI8823, MP3288, DDW47. Analytic measures of adaptability RPGV and MHRPGV pointed out HI8823, MP3288, DBW110 would be more adaptable genotypes.
Biplot analysis of measures
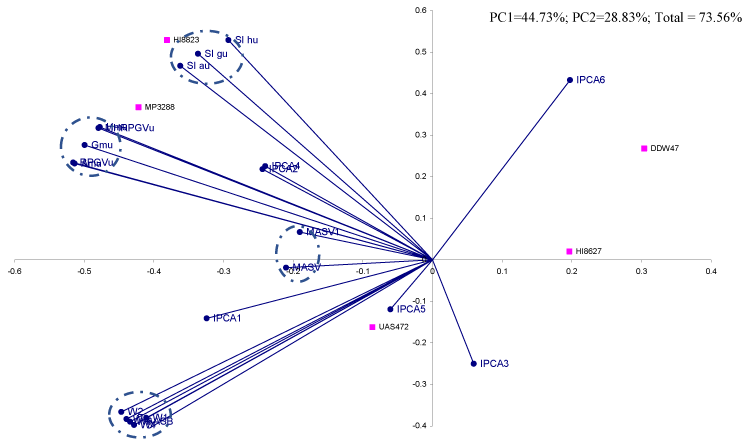
The first two significant PC’s jointly has explained 73.5% of the total variation with 44.7% & 28.8% contributions by PC1 & PC2 (Table 10). The first group comprised of MASV, MASV1 while the second observed for stability measures by utilizing many interaction principal components (Figure 2). Adaptability measures as per arithmetic, geometric and harmonic means along with the corresponding values of RPGV & MHRPGV expressed bondage and placed in a different quadrant. However, this group maintained nearly the right angle with stability measures. The cluster of Superiority indexes as per averages of the yield of wheat genotypes seen in the same quadrant. The performance difference of genotypes would be very minimum by Superiority indexes as compared to adaptability measures.
Conclusions
GxE interaction in multi-loation trials had been studied by AMMI model. Recent analytic measures advocated simultaneous use of stability & yield to recommend high-yielding stable wheat genotypes. Infact both BLUP and AMMI have their efficacy increased depending on factors intrinsic to analysis. In the present study, the main advantages of AMMI and BLUP had been combined to increase the reliability of multi-locations trials analysis. The more interesting advantage provided by Superority Indexes that different weights may be assigned to the yield performance and stability. As per the goal of a breeding program or a cultivar recommendation trial, the researcher may prioritize the productivity of a genotype rather than its stability (and vice-versa). The stability index of genotype performance has the potential to provide reliable estimates of stability in future studies along with a joint interpretation of performance and stability in a biplots while considering more of IPCA’s.
The wheat genotypes were evaluated at research fields at coordinated centers of AICW&BIP across the country. The first author sincerely acknowledges the hard work of all the staff for field evaluation and data recording of wheat genotypes.
- Agahi K, Ahmadi J, Oghan HA, Fotokian MH, Orang SF (2020) Analysis of genotype × environment interaction for seed yield in spring oilseed rape using the AMMI model. Crop Breeding and Applied Biotechnology 20: e26502012. Link: http://bit.ly/3rQ2enh
- Ajay BC, Aravind J, Fiyaz RA, Kumar N, Lal C, et al. (2019) Rectification of modified AMMI stability value (MASV). Indian J Genet 79: 726-731. Link: https://bit.ly/3rPAZcH
- Piepho HP, Mohring J, Melchinger AE, Bu¨chse A (2008) BLUP for phenotypic selection in plant breeding and variety testing. Euphytica 161: 209–228. Link: https://bit.ly/3eRDTtV
- Ashwini KVR, Ramesh S, Sunitha NC (2021) Comparative BLUP, YREM-based performance and AMMI model-based stability of horse gram [Macrotyloma uniflorum (Lam.) Verdc.] genotypes differing in growth habit. Genet Resour Crop Evol 68: 457-461. Link: https://bit.ly/3ljhkiY
- Olivoto T, Lucio A Dal’Col, Gonzalez, Silva JA da, Marchioro VS (2019) Mean performance and stability in multi-environment trials I: Combining features of AMMI and BLUP techniques. Agron J 111: 1-12. Link: https://bit.ly/3timG0u
- Bocianowski J, Niemann J, Nowosad K (2019) Genotype-by environment interaction for seed quality traits in interspecific cross-derived Brassica lines using additive main effects and multiplicative interaction model. Euphytica 215: 1-13. Link: https://bit.ly/3vy8x1l
- Sa’diyah H, Hadi A F (2016) AMMI Model for Yield Estimation in Multi-Environment Trials: A Comparison to BLUP. Agriculture and Agricultural Science Procedia 9: 163-169. Link: http://bit.ly/3rNzBah
- Mohammadi R, Amri A (2008) Comparison of parametric and non-parametric methods for selecting stable and adapted durum wheat genotypes in variable environments. Euphytica 159: 419-432. Link: https://bit.ly/3vszOSs
- Zali H, Farshadfar E, Sabaghpour SH, Karimizadeh R (2012) Evaluation of genotype × environment interaction in chickpea using measures of stability from AMMI model. Ann Biol Res 3:3126–3136. Link: http://bit.ly/3vsif5h
- Resende MDV, Duarte JB (2007) Precision and Quality Control in Variety Trials. Pesquisa Agropecuaria Tropical 37: 182-194. http://bit.ly/3vmBTiW
- Mendes FF, Guimarães LJM, Souza JC, Guimarães PEO, Pacheco CAP, et al. (2012) Adaptability and stability of maize varieties using mixed model methodology. Crop Breeding and Applied Biotechnology 12: 111-117. http://bit.ly/3lt3ZVn
- Gauch HG (2013) A simple protocol for AMMI analysis of yield trials. Crop Sci 53: 1860–1869. Link: https://bit.ly/3lhhtTO
- Ajay BC, Bera SK, Singh AL, Kumar N, Gangadhar K, Kona P (2020) Evaluation of Genotype × Environment Interaction and Yield Stability Analysis in Peanut Under Phosphorus Stress Condition Using Stability Parameters of AMMI Model. Agric Res 9: 477–486. Link: https://bit.ly/3lhK23r
- Oyekunle M, Menkir A, Mani H, Olaoye G, Usman IS, Ado S (2017) Stability analysis of maize cultivars adapted to tropical environments using AMMI analysis. Cereal Res Commun 45: 336–345. Link: https://bit.ly/38Ignvc
- Resende MDV (2007) Software Selegen – REML/BLUP: sistema estatístico e seleção genética computadorizada via modelos lineares mistos. Embrapa Florestas, Colombo, 350.
- Lin CS, Binns MR (1988) A superiority measure of cultivar performance for cultivar x location data. Canadian Journal of Plant Science 68: 193-198. Link: http://bit.ly/3lh8zpq
- Annicchiarico P (1992) Cultivar adaptation and recommendation from alfalfa trials in northern Italy. Journal of Genetics and Plant Breeding 46: 269-278. Link: http://bit.ly/38BVphC
- Mohammadi M, Sharifi P, Karimizadeh R, Jafarby JA, Khanzadeh H, et al. (2015) Stability of grain yield of durum wheat genotypes by AMMI model. Agric For 61: 181-193. Link: https://bit.ly/3eE8CKF
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
