International Journal of Agricultural Science and Food Technology
Clustering and principal component analysis of Barley (Hordeum volugare L.) Landraces for major morphological traits from North Western Ethiopia
Melkamu Enyew1*, Tiegist Dejene2, Berhane Lakew3 and Fisseha Worede4
2Bahir Dar University, Biotechnology Research Institute, Bahir Dar, Ethiopia
3Ethiopian Institute of Agricultural Research, Holleta Research Center, Addis Ababa, Ethiopia
4Ethiopian Institute of Agricultural Research, Fogera Rice Research and Training Center, Bahir Dar, Ethiopia
Cite this as
Enyew M, Dejene T, Lakew B, Worede F (2019) Clustering and principal component analysis of Barley (Hordeum volugare L.) Landraces for major morphological traits from North Western Ethiopia. Int J Agric Sc Food Technol 5(1): 058-063. DOI: 10.17352/2455-815X.000043Huge collections of barley landrace genotypes in Ethiopian are not studied for the magnitude of genetic distances from each other. Though knowing the contribution of individual traits is crucial to focus on particular traits in cultivar development; the traits of these genotypes are not yet studied. Hence, this experiment was conducted on 48 barley landrace accessions which were not studied yet and four standard checks to estimate the magnitude of genetic diversity among the genotypes and to identify the major morphological traits contributing for the observed variations. The experiment was laid in Augmented Randomized Complete Block Design in six blocks at Adet in 2016/17 cropping season. The traits used for analysis were days to 50% heading, days to maturity, plant height, total number of tillers/plant, effective number of tillers/plant, number of spikes/plant, spike length, average number of grains/plant, biomass yield/ha, thousand grain weight/gm, grain yield tones/ha and harvest index tones/ha. The 52 genotypes were grouped into six clusters where 65.39% of the genotypes (34) fall in cluster I, III and IV. Early matured genotypes were grouped in cluster III, late matured in cluster VI and high yielding and tall genotypes in cluster IV. The highest intra cluster distance was 22.513 for cluster III and IV. The highest inter cluster distance was 57.00 between cluster V and IV. The first three principal components contributed 74.20% of the total variations observed among the genotypes. Principal component one (Pc1) alone had contributed 49.96%.% of the total variations mainly due to grain yield, biomass yield, thousand grain weight and plant height in their respective order. Principal component two (Pc2) contributed 15.98% of the total variations mainly through total number of tillers per plant, number of effective tillers per plant and number of spikes per plant in their descending order. Principal component three (Pc3) had contributed 8.25% of the total variations through days to maturity, days to 50% heading and number of grains per plant. The result ensures the existence of high genetic divergence among the studied genotypes.
Introduction
Barley (Hordeum vulgare L.) belongs to the tribe Triticeae and the grass family Poaceae. It is a diploid self-pollinating cereal crop having seven pairs of chromosomes (2n=2x=14) and genome size of about 5.1 GB [1]. Ethiopia is considered as one of the Vaviloves center of diversity for barley endowed with diverse agro-ecologies and wide altitudinal ranges from 110m.b.s.l to 4620m.a.s.l. Long-term mutations, hybridization, gene recombination and natural and human selection in heterogeneous environments inconsistent with edaphic factors create diverse barley landraces in the country. As a result there are over 15 thousand barley landrace accessions collected in the gene bank of Ethiopian biodiversity institute [2]. Though the collections are numerous witwh diverse characters which may be different or inter related, the magnitude of variation is not yet studied. Therefore grouping the existed genotypes based on morphological characters enables breeder to exploit existed genetic resources for further breeding programs. It is real that knowing relationship among the existed landraces enables breeders to develop extreme phenotypes for cultivar development. It is also important to knowing the major traits contributing for the total observed variations among the landraces. This entails breeders to focus on specific characters to develop varieties for specific environmental conditions allowing answering priority community problems. Therefore this study was organized to estimate the magnitude of genetic distance and to identify the major traits contributing for the observed variations among the studied genotypes.
Materials and Methods
Description of the study area
The study was conducted at Adet Agricultural Research Center (AARC). Adet is located 11016’N latitude and 37029’E longitude with an altitude of 2240m.a.s.l. The mean annual rainfall of AARC is 1250mm ranging between 860mm and 1771mm [3]. The average annual maximum temperature of the center is 25.5 °C and the average minimum temperature is 9.2 °C. The soil type of Adet from which this experiment was conducted is red Nitosol with slight clay nature.
Treatments and experimental design
The study was conducted on 48 barley landrace accessions which were not studied yet (obtained from Ethiopian Biodiversity Institute) and four standard checks (Abay, Tilla, Setegn and Mulu gained from Adet Agricultural Research Center).The treatments were coded from one to fifty two in which one up to four corresponds for checks and the rest belonging for the landrace accessions following their ascending order from the passport code. The landraces are indicated in table 1. The experiment was laid in augmented randomized complete block design in six blocks with 2.5m x 4.8m size each. The numbers of blocks were fixed by using the formula applied to determine the minimum number of blocks in augmented block design with respect to the replications of checks. The formula used to determine the number of blocks was as follows [4].
b≥ [(10/r-1)]+1
Where r-number of checks=4, b-number of blocks. b≥ [(10/4-1)]+1, b≥4.3. Accordingly block number for this study should be greater than or equal to five. In this study as number of new treatments were 48 and not equally divisible by five, it was better to make block number as six. The block to block distance was 1m and each block contains eight landraces and four checks. The checks were replicated in each block and the landraces were assigned to plots randomly. The rows were considered as plots with 2.5mx0.4m length and row to row distance respectively. The total experimental area was 4.8m X 20m.
Data collection
In this study both plant based and plot based traits were taken. For plant based traits i.e. plant height, total number of tillers per plant, number of effective tillers per plant, number of spikes per plant, spike length and average number of grains per plant. Five randomly selected plants were tagged at the early stage and measured timely according to the traits used. The averages were used for the analysis. Plot based traits i.e. thousand grain weight, biomass yield and grain yield were taken from the whole plot and converted into hectare bases for the analysis. Harvest index were estimated by dividing grain yield by biomass yield multiplied by hundred. In addition days to 50% heading and days to maturity were counted from emergence to 50% heading and 90% maturity respectively.
Data analysis
The seed for the landrace accessions collected from Ethiopian biodiversity institute were not enough to use replicated designs and it was obligatory to use augmented designs in such circumstances. As a result the checks were replicated in each block and the landraces were not. The row data were adjusted to mean of zero and variance of one by using the means of checks in each block and the overall mean of checks in the whole plot. This was to minimize errors brought due to unreplicated treatments. The formula used for the data adjustment was as follows [4].
ýij =yij –x-i –x=
where ýij –adjusted mean of each observation, yij –original observation of each genotype, x-i –mean of checks in each block and x= - grand mean of checks in all blocks.
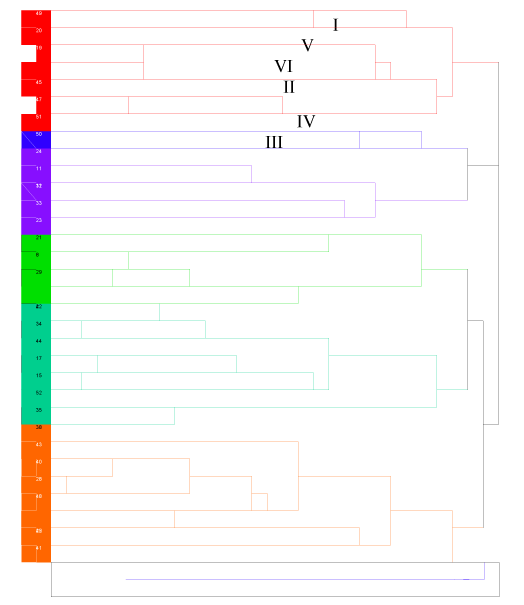
Cluster analysis: The 52 genotypes were clustered into six by hierarchical clustering with average linkage method, using standardized values of 12 traits at mean of zero and variance of one by SAS 2008 (version 9.2) software. The numbers of clusters were decided by using pseudo f and t-test. The distance between and within clusters were also estimated. Finally, clustered genotypes were displayed with dendrogram in different colors.
Principal component analysis (Pca): It is important to estimate the contribution of each trait for the total observed variations in the genotypes. This enables to identify the major traits accounting the greater share in the observed variations. This enables breeder to focus on specific traits of interest for crop improvement. Therefore principal component analysis is important to estimate the contribution of each trait in percent bases. The analysis was computed by using SAS 2008 (version 9.2) up to twelve principal components. The first four principal components for the 12 characters were taken as they account the highest share for the total variations. The values were taken interns of Eigen vectors and Eigen values from correlation matrix. The genotypes were categorized in bi-plot figure and compared with the cluster analysis.
Results and Discussion
Cluster analysis
The data were standardized to the mean of zero and variance of one with SAS 2008 prior to analysis. Then, the 52 genotypes were clustered hierarchically with average linkage clustering method into six clusters. In the analysis 12 genotypes (23.08%) were grouped in cluster IV, 12 genotypes (23.08%) in cluster III, seven genotypes (13.46%) in VI, ten genotypes (19.23%) in cluster I, three genotypes (5.77%) in cluster V and the rest seven genotypes (13.46%) into cluster II. The analysis showed that the genotypes were distributed in different clusters indicating existence of variations among the studied genotypes, showing high probability of recombination. The lists of genotypes in each cluster are displayed in table 2.
In consistent with this result Bedasa Mekonnon et al., [1], studied other 102 Ethiopian barley landraces accessions and five standard checks (this landraces are not parts of this study) and grouped these genotypes into five clusters. They found maximum genotype (44 genotypes) in cluster I and minimum genotypes (two genotypes) in cluster V. The rest of the genotypes were distributed in the remaining clusters ensuring the availability of genotypic variability among the landraces.
Likewise Zeynu Tahar et al., [5], studied other 36 Ethiopian barley genotypes and the report indicated that the genotypes were clustered into seven clusters. The number of genotypes in the report ranged from two in cluster I & III to eight genotypes in cluster IV. They also stated that the distribution pattern of the genotypes into various clusters showed the presence of considerable genetic divergence among the genotypes for days to heading, days to maturity, plant height, spike length, number of productive tillers per plant, number of kernels per spike, biomass yield per hectare, harvest index and thousand grain weight.
Addisu Fekadu et al., [6], also studied 36 barley landrace accessions in southern Ethiopia and categorized them into four clusters. According to their study the maximum numbers of genotypes (11) were grouped in cluster II and IV and the minimum number were four in cluster II. These studies confirm that Ethiopian barley landraces have great genetic variability.
Cluster III contains on average base early mature (79.64 days), moderately yielding (0.82 tones/ ha), medium height (93.95cm) landraces. Cluster IV contains high yielding (1tones/ha), high thousand grain weight (34.85gm), moderate maturity date (93.02 days), tall plant height (100.45cm) and medium number of grains (23.17 grains) per plant. Cluster II contains landraces that are moderately yielding (0.73tones/ha), high number of grain (36.41 grains) per plant and high harvest index (0.31%). Cluster VI contains landrace that are late mature (108.68 days), low yielding (0.20 tones/ha) and low harvesting index (0.16%). Whereas the genotypes grouped in cluster I and V performed lower than genotypes in other clusters.
In agreement with the results in this study Zeynu Tahar et al., [5], reported that genotypes which are early heading ( 66.1 days), have high number productive tillers and less amount of kernel number per spike were grouped in cluster I. Genotypes with small number of productive tillers, grain yield and thousand kernel weight were grouped in cluster II. He also reported that genotypes with tall plant height (111.2cm) were grouped in cluster V.
Similarly a study by Kemelew Muhe and Alemayehu Assefa [7], on another 181 Ethiopian barley landraces had grouped the genotypes into ten clusters. The report showed that high yielding (2.634, 1.898, 2.242, 2.063 and 1.728 tones/ha) genotypes were grouped in cluster I, II, III, IX and X in their respective order. They also indicated that landraces with low yielding (1.28, 1.05, 0.833 and 0.662 tones/ha) and low thousand grain weights (37.12, 36.09, 33.93, 33.20gm) were grouped in VI, IV, VIII and VII respectively.
A study conducted on another 102 Ethiopian barley landrace accessions by Bedasa Mekonnon1 et al., [1], also indicated that genotypes with high plant height (106.53cm) on average bases grouped in cluster II. The report showed that the mean thousand seed weight of the clusters ranged from 46.62, 43.06, 46.96 and 42.30 for cluster I, II, III and IV respectively confirming existence of genotyping variability. According to the study the highest mean cluster value for 50% days to heading and maturity were 84.12 and 130.34 accordingly in cluster IV (Table 3).
DF-Days to 50% flowering, DM-Days to Maturity, PH- Plant Height/cm, TTN/plant-Total Tiller Number Per Plant, ETN/plant-Effective Tiller Number Per Plant, NS/plant-Number of Spikes Per Plant, SL-Spike Length(cm), NG/plant- Average Number of Grains Per Plant, BM-Biomass Yield(t/ha), Thousand Grain Weight(gm), GY– Grain Yield(t/ha) and HI- harvest index on hectare bases.
On average the highest intra cluster distance (distance within clusters) was 22.51for cluster III and IV. This indicated that genotypes in the same cluster (in cluster III and IV) had sufficient distances with each other for recombination in cultivar development. The lowest intra cluster distance was 3.2475 for genotypes within cluster V indicating the genotypes within this cluster are more similar.
The highest inter cluster distances were 59.5163 between clusters III & VI, 56.9996 between cluster IV and V; and 54.7799 between Cluster IV and VI. The lowest inter cluster distances were 17.6859 between cluster III and IV; 26.6721 between cluster II and IV; and 30.0017 between cluster II and III. The results of inter cluster analysis revealed that the genotypes had wide genetic divergence with each other indicating existence of high probability for recombination. Results from cluster analysis also strengthen the availability of genotypic variability even within clusters.
In line with this finding Zeynu Tahir reported that the highest inter cluster distance as 205.82 between clusters I & V and the lowest inter cluster distance as 20.43 between cluster VII and VIII. He confirmed that there is the existence of wide genetic divergence among the landraces expected to manifest maximum heterosis in crossing and wide genetic variability.
Align with this study Shegaw Derbew et al., [8], also studied on 225 landraces and reported that highest inter cluster distance as 47.0 between Cluster III and VIII and as 42.4 between cluster VII and IX. The study indicated that the lowest inter cluster distance was 12.3 between cluster V and II (Table 4).
The dendrogram assures that the highest numbers of genotypes (12 in each cluster) were grouped in cluster III and IV and the lowest number of genotypes (3) were grouped in cluster V. It also revealed that the genotypes were classified into two wider groups’ group one containing cluster I, V & VI and the other group containing cluster II, III &IV. Genotypes within these two wider groups indicate that there is a wide variability among genotypes between the wider clusters to undertake crossing. The genotypes within cluster IV and cluster III were classified into many sub categories showing availability of high variability with in clusters. The classifications within cluster V are small as compared to the rest of the clusters indicating availability of low genotype variability within this cluster. The dendrogram indicated that the lowest inter cluster distance were between cluster III and IV which are putted side by side. The highest was between cluster III and IV. The dendrogram result ensures that the graphical display is consistent with estimated numerical values within and among clusters.
Genotypes within the same cluster are marked with similar color and genotypes from different cluster are distinguished with different color. Closely related genotypes are marked with similar symbol prior to the treatment code in the graph. The numbers in the graph shows genotype codes each corresponding to accessions which were described in table 2.1 above. Example 1 is for C-1(Abay), 2 is for C-2 (Mulu), 3 is for C-3 (Setegn), 4 is for C-4(Tilla), ………… 52 is for accession IC-223953.The genotypes with their represented codes are displayed in figure 1.
Principal component analysis
The data of 52 barley genotypes were prepared as correlation matrix for the 12 traits (12*52) and analyzed by SAS 2008 (version 9.2) software. The first four principal components that had better contributions for the observed variations were taken from the 12 principal components. These principal components had contributed 80.79% of the total variation for the 12 characters under study. Principal component one (PC1) alone had contributed 49.96% of the total variation. For principal component one grain yield, biomass yield, thousand grain weight and plant height had contributed 0.3679, 0.3494, 0.3352 and 0.3156 in their respective order. Total number of tillers per plant had low contribution for the observed variations in PC1. However days to 50% heading and days to maturity had no contribution for PC1 rather they counter balance the observed variations. There for most of the variations among genotypes in principal component one was brought due to the four major traits indicated above.
Principal component two (PC2) had contributed 15.98% of the total variation. The observed variations in PC2 were mainly brought by total number of tillers per plant, number of effective tillers per plant and number of spikes per plant contributing 0.6277, 0.4464 and 0.393 in their descending order. Principal component three (PC3) accounts 8.25% share of the total variations which were mainly contributed by days to maturity, number of grains per plant and days to 50% heading accounting 0.6578, 0.4997 and 0.4242 in their respective order. Principal Component four (PC4) accounts 6.61% of the total variations observed among the studied genotypes for the 12 characters. The variations for PC4 were majorly brought by grain yield, days to maturity, harvest index and biomass yield sharing 0.3984, 0.3322, 0.3248 and 0.3113 respectively.
In agreement with this study Seid Ebrahim et al., [9], studied other 20 Ethiopian barley varieties from Wollo highlands and reported that the first three principal components together exerted 84.22% of the total variations observed among the barley genotypes through kernel number per spike, kernel weight per spike, number of spikelet per spike, thousand seed weight, spike length, grain yield per plot, days to heading and maturity.
A study by Kiflu Tarekegn [10], on another 20 Ethiopian barley landraces also reported that the first three principal components contributed 73.14% of the total variations contributed through productive tillers per plant, grain yield and thousand grain weight, spike length and awn length from the total genotypic variations observed (Table 5).
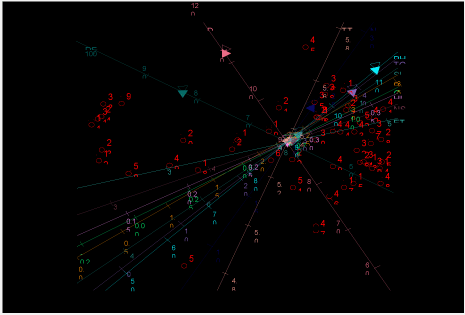
The biplot graph below in figure 2 also indicated the variability of genotypes for the 12 traits under study. The graph was plotted by scaling each trait independently to separate genotypes. Those genotypes close to the center are more closely related with each other and the distant genotypes are more divergent. Accordingly genotype 5 and 45 are more divergent with each other for total tiller number per plant and number of grains per plant. Genotype 19, 22, 30 and 6 are more close to the center of the biplot graph indicating less divergence for the studied 12 traits. The genotype 33, 9, 31, 24 and 11 had high distances from genotypes 48, 25, 14, 28 and 41 indicating the possible distant genotypes for crossing.
DF-Days to 50% flowering, DM-Days to Maturity, PH- Plant Height(cm), TTN/plant-Total Tiller Number Per Plant, ETN/plant-Effective Tiller Number Per Plant, NS/plant-Number of Spikes Per Plant, SL-Spike Length(cm), NG/plant- Average Number of Grains Per Plant, BM-Biomass Yield(t/ha), Thousand Grain Weight(gm), GY– Grain Yield(t/ha) and HI- harvest index on hectare bases [11-13].
Conclusion
Ethiopia is endowed with hug barley landraces genetic resources enabling it as the centers of origin and diversity. These large collections of barley landraces are divergent and rich with various unique traits. However the magnitude of these genetic divergences is not quantified yet. As a result the country is not befitted from its own genetic resources. Therefore this study was organized to estimate the genetic distance among the studied genotypes. The studied 52 genotypes were distributed into six clusters. On average the maximum genetic distances among the studied barley landraces were 59.5163. The result ensures that there is a high genetic divergence among the studied genotypes. The major traits contributing for these observed variations among the studied genotypes were grain yield, biomass yield; thousands grain weight and plant height contributing 49.96%. As a result the landraces could be used as contrasting parents for further breeding programs. Yield of these landraces could be improved by either crossing these divergent landraces with each other or by selection focusing on major traits.
- Mekonnon B (2015) Morphological diversity and association of traits in Ethiopian food barley (Hordeum volgare L.) landraces in relation to regions of origin and altitudes. Journal of Plant Breeding and Crop Science 7: 243-253. Link: http://bit.ly/2YbSo4q
- Engels (1994) Genetic diversity in Ethiopian barley in relation to altitude. Genetic Resource and Crop Evolution 41: 67-73. Link: http://bit.ly/2Yvo545
- Bayeh B (2010) Assessment of bread wheat production, marketing and selection of N- efficient bread wheat (Tritium aestivum L.) varieties for higher grain yield and quality in North Western Ethiopia. MSc. thesis, Bahir Dar University, Ehiopia. Link: http://bit.ly/331xNhT
- Federer WT, Raghavarao D (1975) On Augmented Designs. International Biometric Society 31: 29-35.
- Tahar Z (2016) Phenotypic diversity in Ethiopian food barley (Hordeum vogare L.) genotypes. World Journal of Agricultural Sciences 12: 308-314. Link: http://bit.ly/2ZplMAW
- Fekadu A (2018) Qualitative traits variation in Barley (Hordeum Vulgare L.) landraces from Southern Highlands of Ethiopia. International Journal of Biodiversity and Conservation 10: 258-264. Link: http://bit.ly/2YhmAem
- Kemelew, Assefa A (2011) Diversity and Agronomic Potential of Barley (Hordeum Vulgare L.) landraces in Variable Production Systems, Ethiopia. World Journal of Agricultural Science 7: 599-603. Link: http://bit.ly/2SZETz6
- Derbew S (2013) Genetic Variability in Barley (Hordeum Vulgare L.) Landrace Collections from Southern Ethiopia. International Journal of Science and Research 2319-7064.
- Ebrahim S (2015) Evaluation of genetic diversity in barley (Hordeum volgare L.) from Wollo highland areas using agro-morphological traits and hodein.” African Journal of Biotechnology 14: 1886-1896. Link: http://bit.ly/331YZxi
- Tarekegn K (2009) Agronomic evaluation of Ethiopian barley (Hordeum Vulgare L.) landrace populations under drought stress conditions in low rain fall Areas.” MSc. Thesis No.61, International Master Program at Swedish Biodiversity Center, Uppsala University, Sweden. Link: http://bit.ly/2MBQR0L
- Akintunde A (2012) Path analysis step by step using excel. Journal of Technical Science and Technologies 1: 9-15. Link: http://bit.ly/2Kg5WlK
- Ali MK (2014) Investigate the relationship between studied traits with grain yield using regression analysis and path analysis in 34 barley lines and cultivars. Science and Research Tehran Branch 4: 42-45. Link: http://bit.ly/2Ywtzzp
- SAS institute Inc (2008) Statistical Analysis Software version 9.2, USA, Cary, NC: SAS Institute Inc. Link: http://bit.ly/2KhfigR
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
