International Journal of Aquaculture and Fishery Sciences
Nidoviruses in Aquatic Organisms - Paradigm of a Nascent Concern
Monalisha Kumar, R. Bharathi Rathinam and Gayatri Tripathi*
Cite this as
Kumar M, Rathinam RB, Tripathi G (2020) Nidoviruses in Aquatic Organisms - Paradigm of a Nascent Concern. Int J Aquac Fish Sci 6(3): 082-088. DOI: 10.17352/2455-8400.000061The extraordinarily genetically complex Nidovirus is an enveloped, single-stranded RNA virus. This diversified group of viruses has a higher degree of RNA recombination due to its large nested arrangements of sub-genomic mRNA. This is considered to be crucial for accelerating the evolutionary process and pathogenesis of the virus resulting in enhancing their adaptation to rapidly changing environment as well as upgrading them to infect a broad range of host. In general, Nidoviruses are an alarming group of pathogens of terrestrial animals but they have, of late, started affecting a wide range of aquatic animals such as fish and shellfish since the last 20 years. The common Nidoviruses reported from aquatic animals are Bafinivirus i.e. White bream virus, Fathead minnow virus, Chinook salmon bafinivirus from fish; Yellow head virus, Gill-associated virus from shrimp; Eriocheir sinensis ronivirus from crab; Harbour seal coronavirus, Beluga whale coronavirus, Bottlenose dolphin coronavirus from marine mammals. The clinical symptoms associated with this virus infection are mainly noticed in the skin, eye, anterior kidney, spleen and liver whereas gill, lymphoid organ, haemopoietic tissue, midgut, cuticular epithelium, heart, haemocytes in shrimps and ecdysal gland in crabs are the primary target organs of Nidoviruses. It has gained considerable importance in the research area of aquatic animal health with the discovery of first fish Nidovirus in cyprinids but the prevalence, host range, route of infection, mode of transmission and diagnosis of the virus are required to be explored in detail to evaluate the significant risk of Nidoviruses and to control the future outbreak of diseases in aquatic animals.
Abbreviations
ORF: Open Reading Frame; nt: Nucleotides; WBV: White Bream Virus; FHMNV: Fathead Minnow Nidovirus; PCR: Polymerase Chain Reaction; RT-LAMP: Reverse Transcriptase-Loop Mediated Isothermal Amplification; CCNV: Crucian Carp Nidovirus; CSNV: Chinook Salmon Nidovirus; YHV: Yellow Head Virus; GAV: Gill-Associated Virus; EsRNV: Eriocheir sinensis Ronivirus; BWCoV: Beluga Whale Coronavirus; BdCoV: Bottlenose dolphin Coronavirus; CoV: Coronavirus; SARS: Severe Acute Respiratory Syndrome; MERS: Middle East Respiratory Syndrome
Introduction
The Order “Nidovirales” [1] was introduced for the first time by the International Committee on Taxonomy of Viruses in the year 1996. The Latin term ‘nidus’ means nests inferring the nested assemblage of sub-genomic mRNA [2]. Nidoviruses possess a wide variety of morphological and molecular structures which justify assigning these viruses into different families. The intricate molecular genetics completely discriminating the aquatic nidoviruses from other groups of RNA virus which enables them to survive in different ecological niches. Group IV positive-sense single-strand RNA nidovirus is grouped under two categories based on their genome size; large nidovirus and small nidovirus. Large nidovirus comprises three families Coronaviridae, Roniviridae, Mesoniviridae but the small nidovirus consists only a single family, viz., Arteriviridae [3-5]. The nidoviruses can infect a wide range of hosts and can acclimatize to adverse environmental conditions due to their higher frequency of recombination and mutation [6]. As a result, the taxonomy of nidovirus is constantly changing. Thus, the establishment of complex replication-transcription model of all nidoviruses has been one of the thriving areas of research. It is projected to be a progressive field of study with the commencement of Nidoviruses in aquatic animals after breaking down the perception that Nidoviruses are restricted to terrestrial animals i.e. particularly humans and livestock animals. It affects a broad range of hosts from marine mammals to fishes as well as shrimps and crabs. Among these, Bafinivirus infects the fish population whereas crustaceans are affected by Okavirus and aquatic mammals by AlphaCoronavirus and GammaCoronavirus Table 1.
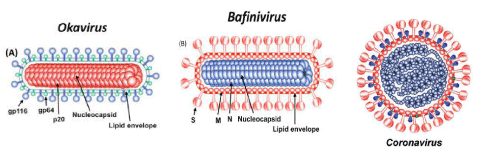
Virion morphology and genome organization
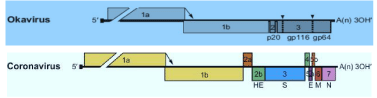
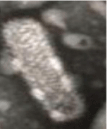
The aquatic nidoviruses are enveloped, bacilliform (Bafinivirus and Okavirus) or spherical shaped (Coronavirus) viruses containing positive sense single strand, non-segmented, linear RNA genome. The capped and polyadenylated RNA genome size is 27-33 kb. The average diameter is 120 to 160 nm for Coronaviridae family and 45 nm for Roniviridae family whereas mean relative molecular mass (Mr) - 400 × 106 and buoyant density (Bd) in sucrose is 1.13–1.20 g/cm3 [3]. In general, ORF 1b is the most conserved part associated with transcription for synthesizing RNA to produce the new virions in nidovirus. A distinct set of genetic markers - a putative multinuclear zinc-binding domain (ZBD) and a uridylate-specific endoribonuclease (NendoU) domain are found in nidoviruses which play a vital role in the replication process [6]. ORF1a and ORF1b are translated by -1 ribosomal frameshifting. The resultant polyproteins pp1a and pp1ab produce the viral RNA dependent RNA polymerase (RdRp) and other non-structural proteins involved in replication-transcription event. The Receptor-Binding Domain (RBD) present in the spike protein is the most variable part of this viral genome. Structural proteins such as spike glycoprotein, envelope protein, membrane protein, nucleocapsid protein are expressed as sub-genomic RNAs and essential to regulate the entry of pathogen to host cell, form a viral envelope, determine the shape of the envelope, make up the nucleocapsid as well as organize the viral assembly etc [21] Figures 1,2.
Life-cycle
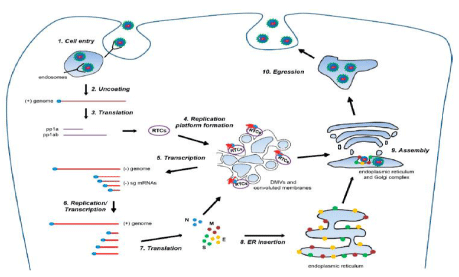
Infection begins when ligand of nidovirus particle attaches to a host cell surface receptor and subsequent cell entry occurs via endosomes by the process of endocytosis. Consequently, the genomic RNA of the virus is released in the host cell and translation takes place, which produce the replicase polyproteins i.e., polyprotein 1a (pp1a) and polyprotein 1ab (pp1ab). Further, non-structural proteins are produced through auto-proteolysis followed by the generation of replication-transcription complexes (RTCs). The RTCs are double membrane vesicles (DMVs) with convoluted membranes. At first, minus-strand of RNA are synthesized to yield both full length single strand and sub-genomic minus-strand RNAs with the involvement of RTC complexes. Eventually, the sub-genomic mRNAs are used as templates to form the genomic RNA and plus-strand sub-genomic mRNAs are essential to express the structural protein genes. Structural proteins like spike, envelope, and membrane proteins are synthesized and incorporated in the endoplasmic reticulum whereas the translation and oligomerization of nucleocapsid proteins takes place in the cytosol. Subsequently, the ribonucleoprotein complexes are produced due to the interaction with RTCs and association with the genomic RNA. Virions are assembled in the ER and/or Golgi followed by the inward budding of the limiting membrane of these compartments which is stimulated by the interaction between the ribonucleoprotein complexes and structural proteins. At last, egression takes place where mature virions are coming out of the cell by exocytosis [22]. The reasons behind the complexity of nidovirus transcription are the production of numerous sub-genomic RNAs due to discontinuous mode of transcription during synthesis of minus strand of RNA and the large size of the genome [23] Figure 3.
Nidoviruses in fish
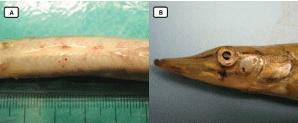
White bream virus (WBV): The virus was first characterized from a healthy white bream or silver bream in Germany [7]. It encompasses 26.6 kb long genome consisting of five ORFs. It encodes non- structural protein i.e. replicase polyprotein and structural proteins like spike protein, membrane protein and nucleocapsid protein. This virus has been also isolated from other cyprinids like goldfish (Carassius auratus auratus), tench (Tinca tinca) and grass carp (Ctenopharyngodon idella) [24]. As it is not showing detrimental effects on fish; listlessness, anorexia, excess mucus production, pseudofaeces and petechiae are common nonspecific clinical signs [25].
Fathead minnow virus (FHMNV)
In 1997, Fathead Minnow Virus was first reported from moribund fathead minnows (Pimephales promelas) in United States [9]. The viral genome consist of approximately 27 kb RNA and the structural organization of genome resembles to WBV genome. The fish infected with FHMNV exhibited skin haemorrhage, eye haemorrhage, gill palor, multifocal petechial haemorrhages on the abdomen and lesions in liver, spleen, kidney. Fishes challenged with this pathogen suffered 90% mortality. Histopathology is exemplified by nuclear pyknosis displaying marginated chromatin and severe multifocal areas of necrosis in the anterior kidney, liver and spleen [9]. In the study of experimental infection, morbidity becomes apparent by changes in behaviour and appearance characterized by initial erratic swimming followed by listlessness at the tank bottom and head faced upward at the water surface [9]. It had also been detected in other fishes such as muskellunge (closely related to northern pike) [26], golden shiner and spotfin shiner [27]. The diseased spotfin shiner exhibits moderate echymotic haemorrhage on the isthmus and severe multifocal echymotic haemorrhage along the caudal to the dorsal fin whereas golden shiner infected with FHMNV showed severe diffuse haemorrhage in brain and severe multifocal pinpoint haemorrhage along the spine just below the dorsal fin [28].
For diagnosis and surveillance of this pathogen, the development of PCR procedure for FHMNV was accomplished in 2012 [10]. Subsequently, RT-LAMP test with a sensitivity of around 5 copies of RNA [29] and reverse transcriptase PCR assay [26] were standardized for precise diagnosis.
In 2019, Xiao-yu, et al. [30], isolated a nidovirus from crucian carp and named it as Crucian carp nidovirus (CCNV) having the highest resemblance with Chinook salmon nidovirus (CSNV). Another novel nidovirus was reported from cultured chinook salmon [31], posing a threat to the wild stock of salmon as well as in aquaculture Figures 4,5.
Crustacean nidoviruses
Yellow head virus (YHV): Yellow head disease (YHV genotype 1), among the eight known genotypes of the yellow head complex of viruses, was first identified to be associated with the mass mortality in cultured Penaeus monodon in Thailand in 1990 [32]. During 1992-1993, yellow head disease left a devastating effect over shrimp farming and caused an economic loss of over 40 million U.S. dollars [33]. Subsequently, the disease spread over other Asian countries popular for shrimp farming [34]. It’s genome consists of four long ORFs with 26,662 nt. In general, juvenile to subadult P. monodon were more susceptible to infection. Due to the bleached appearance of the infected shrimp as well as the yellowish cephalothorax this disease is referred to as Yellow Head Disease. Yellowish cephalothorax is developed from the combination of brownish gills and swollen yellowish hepatopancreas [32,35]. Ectodermal and mesodermal organ like gill lamellae, lymphoid organ, haematopoietic tissue, connective tissue of the subcutis, gut, antennal gland, gonads, nerve tract, ganglia and haemocytes are the primary target organs for the infection. Histopathology is indicated by marked cellular necrosis, pyknosis and karyorrhexis of nuclei with perinuclear inclusion bodies in lymphoid organ and gill lamellae [36].
Gill-associated virus (GAV): YHV genotype 2, a part YHV complex is commonly known as “Gill-Associated Virus” (GAV). It was first detected in wild and farmed giant tiger shrimp in Queensland, Australia in 1994 [16]. The virus could be found over the whole life-span of wild shrimp either in a dormant or chronic infection state [16,37]. GAV and YHV are two different kinds of virus because they show approximately 80-85% homology in DNA sequence and 96% homology in amino acid composition [38,39]. Reddening of body and appendages, biofouling with ectoparasites, emaciation, pink to yellow discolouration of the gills are important gross pathological signs recorded.
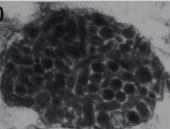
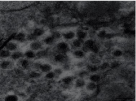
Eriocheir sinensis ronivirus (EsRNV): It causes an infection called, “sighs” disease (SD) in Chinese mitten crabs displaying respiratory problems. The crabs manifested by viruses respire bubbles that sound like ‘sigh’ in the serene of night. It is associated with black gill syndrome in freshwater Chinese mitten crab. Anorexia and sluggishness are non-specific clinical symptoms. The enveloped rod-shaped virus possesses approximately 22 kb long ssRNA genome. There is a close evolutionary relationship between EsRNV and Okaviruses based on structure and genome characteristics but the whole genome sequencing is required to assign this new virus either to an existing genus or to a new genus [40]. The disease damaged over 28,000 hectares land in China causing a huge loss of about 30 lakh euros to the economy in 2002. Histopathology is marked by 1–5 μm diameter clusters of pale to deeply basophilic necrotic cells exhibiting nuclear hypertrophy, karyorrhexis or pyknosis and by 200–800 nm diameter cytoplasmic inclusion bodies. Necrotic cells were observed in the lymphoid organ and connective tissues of different organs such as hepatopancreas, gut, gills, heart and testes [17,41] Figures 6-8.
Crab Oka-Like Viruses: The rod-shaped Rhabdolike-virus A (RhVA) was isolated from the Atlantic blue crab (Callinectes sapidus) in 1970s [43,44]. The infection of this virus was spotted in Y organ or mandibular glands (also known as ecdysial gland). Therefore, later it was renamed as ecdysial gland virus 2 (EGV-2) [45,46]. Soon after, it was noticed that EGV-2 seen to be present along with EGV-1, another rod-shaped viral particle ornamented with peplomers [45]. It was apparent that EGV-1 and EGV-2 appeared conspicuously after the eyestalk ablation. Therefore, it is indicating that disease occurrence is due to stress response to the pre-existing infections [45]. As the diameter of the documented Okaviruses [40] and Bafiniviruses [3] are greater than the rod-shaped RhVA and EGV-2 virions [3], complete genome sequencing is needed to establish their evolutionary relationships.
Nidoviruses in aquatic mammals
Harbor seal Coronavirus: Acute necrotizing enteritis had occurred in the Pacific harbor seal (Phoca vitulina) due to suspected Coronavirus infection [18]. It is a distinct form of AlphaCoronavirus but most closely related to the CoVs of ferrets, cats, dogs, and swine. Markedly heavy, wet, diffusely magenta to purple lungs excluding moderate frothy pink serous fluid from the cut surfaces, moderate serosal petechiation in the small intestine, mucosal linings and white firm multi-cystic mass occupying a part of the left ovary was recorded after diagnosis. Depleted germinal centre and presence of multinucleated syncytial cells were found in the spleen, tonsil affected by this Coronavirus [47].
Beluga whale Coronavirus (BWCoV): A distinct GammaCoronavirus was isolated from a beluga whale by using a pan-viral DNA microarray assay (Virochip). The viral particles were detected in the liver through electron microscopy. The genome (approx. 31.7 kb) comprises 14 putative ORFs encoding eight putative accessory proteins. 5 major ORFs which is known to be conserved in all reported Coronavirus are found in this virus [19].
Bottlenose dolphin Coronavirus (BdCoV): A novel aquatic mammal GammaCoronavirus, Bottlenose dolphin Coronavirus was discovered in 2014. Both Cetacean Coronavirus BdCoV and BWCoV share nearly identical genome organization as per the data of whole genome sequencing and comparative genomics study. But a major distinguishing feature between these two viruses was observed in their structures and composition of spike glycoproteins. The largest genome among all Coronaviruses were detected in BdCoV due to multiple distinct ORFs, the estimated size being around 32000 nt. It is assumed that BdCoV and BWCoV had evolved around 60 years back from a common ancestor of Coronavirus [20] Figure 9.
The COVID-19 pandemic has created a global crisis affecting the world economy [49], its deadly nature is similar to SARS Coronavirus outbreak in 2002 [50] and MERS Coronavirus outbreak in 2013 [51]. In all cases, bats are considered as reservoir hosts of CoVs because of its ability to migrate over extensive areas due to its strength of flight. Due to several recombinations in various CoVs, new viral strains can emerge. Consequently, there will be a tremendous possibility of migrating the pathogens from bats to other animals resulting in the adaptation of new host followed by the transmission to livestock animals or humans [52] (Table 2).
Major difference between Human Coronavirus and Aquatic Coronavirus
Zoonotic diseases are coupled with rapidly changing environment and human behaviour. To date, nidoviruses of aquatic organisms have not been reported to have any zoonotic potential nor has fisheries proven to be associated with the transmission of Coronavirus to humans worldwide in any report. Bondad-reantaso, et al. [53] also stated that SARS-CoV-2 might not infect aquatic animals. Therefore, the fish and fishery products consumption are safe as per the statement given by Department of Fisheries, Ministry of Fisheries, Animal Husbandry and Dairying in 2020 [54]. However, there are chances of transmission of CoV to fish and fishery product, if an infected person handles these products in processing plant or at wet market. The wet markets make an entire squall for cross-species transmission of pathogens. The nidoviruses are suspected to remain in the latent state in many animals [55,56]. The present point of view is that it is necessary to stop the propagation of the virus and forthcoming “zoonotic spill overs”. Different CoVs with zoonotic importance are moving around the wild population but Bat CoVs are the most diverse group. There are ample chances of evolution and recombination of zoonotic CoVs leading to the burgeoning of new CoVs strain which could be more communicable and fatal in humans in near future. Though SARS-CoV-2 have an animal source, it has been noted that the principal route of transmission of Coronavirus seems to be human interaction as per the World Animal Health Organisation (OIE) [57]. The implementation of interdisciplinary ‘One Health’ concept representing a holistic approach including human, animal and environment is the need of the day.
Conclusion
Hitherto, fish Nidoviruses are most prevalent in cyprinids and salmonids. The most common Nidovirus FHMNV severely affects the economically important freshwater baitfish industry in North America. It can also infect hatcheries where baitfish are used as a source of feed to fulfill the protein requirement of farmed fishes. Therefore, some rigorous biocontrol measures such as quarantine, screening and certification are required to control the transfer of infectious pathogen FHMNV to other farmed animals. Roniviridae, genus Okavirus (YHV) causes substantial economic losses in crustacean farming, therefore, the World Organization for Animal Health has declared it as an OIE notifiable disease. The systematic surveillance of aquatic mammal populations maintained in zoos and aquaria is crucial for ensuring the public health safety. Simultaneously, the discovery of new nidoviruses through continuous screening from the wild and cultured aquatic animals will aid to understand the mechanism of mutation and recombination in the viral genome. Such investigations will provide understanding of the evolutionary biology of Nidoviruses and their pathogenesis. Furthermore, the health of humans is intricately connected with the health of animals and the environment. Thus, a holistic approach, called “One Health” needs to be developed taking into account the global health issues, climate change, migration and its interconnections. Hence, it is important to understand the need of “One Health” concept and work on all the associated aspects to protect the health of humans, animals, and the environment.
- Pringle CR (1996) Virus taxonomy 1996—a bulletin from the Xth International Congress of Virology in Jerusalem. Arch Virol 141: 2251-2256. Link: https://bit.ly/34fI65g
- Cavanagh D (1997) Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol 142: 629-633. Link: https://bit.ly/2YwcLHV
- De Groot RJ, Cowley JA, Enjuanes L, Faaberg KS, Perlman S, et al. (2012) Order nidovirales. In: King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J. (Eds.), Virus Taxonomy, Classification and Nomenclature of Viruses. Ninth Report of the International Committee on Taxonomy of Viruses Elsevier Academic Press, San Diego, CA 785-795.
- Lauber C, Ziebuhr J, Junglen S, Drosten C, Zirkel F, et al. (2012) Mesoniviridae: a proposed new family in the order Nidovirales formed by a single species of mosquito-borne viruses. Arch Virol 157: 1623–1628. Link: https://bit.ly/2E7MyYT
- Nga PT, Parquet MD, Lauber C, Parida M, Nabeshima T, et al. (2011) Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes. PLoS Pathog 7: e1002215. Link: https://bit.ly/3gflLHc
- Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ (2006) Nidovirales: evolving the largest RNA virus genome. Virus Res 117: 17-37. Link: https://bit.ly/2CE6t0S
- Granzow H, Weiland F, Fichtner D, Schütze H, Karger A, et al. (2001) Identification and ultrastructural characterization of a novel virus from fish. J Gen Virol 82: 2849-2859. Link: https://bit.ly/324GKaO
- Schütze H, Ulferts R, Schelle B, Bayer S, Granzow H, Hoffmann B, et al. (2006) Characterization of White bream virus reveals a novel genetic cluster of nidoviruses. J Virol 80: 11598-11609. Link: https://bit.ly/34fIAs6
- Iwanowicz LR, Goodwin AE (2002) A new bacilliform fathead minnow rhabdovirus that produces syncytia in tissue culture. Arch Virol 147: 899-915. Link: https://bit.ly/2Q7kEyF
- Batts WN, Goodwin AE, Winton JR (2012) Genetic analysis of a novel nidovirus from fathead minnows. J Gen Virol 93: 1247-1252. Link: https://bit.ly/2FwL1fm
- Lord SD, Raymond MJ, Krell PJ, Kropinski AM, Stevenson RM (2014) Novel chinook salmon Bafinivirus isolation from ontario fish health monitoring. In Proceedings of the seventh international symposium on aquatic animal health, Portland, Oregon, United States.
- Jiang Y, Thomsen I (1987) A new virus isolated from cultured grass carp Ctenopharyngodon idella. Dis Aquat Org 3: 181-185.
- Sano T, Yamaki T, Fukuda H (1988) A novel carp Coronavirus, characterization and pathogenicity. International Fish Health Conference, Vancouver, Canada.
- Miyazaki T, Okamoto H, Kageyama T, Kobayashi T (2000) Viremia-associated ana-aki-byo, a new viral disease in color carp Cyprinus carpio in Japan. Dis Aquat Organ 39: 183-192. Link: https://bit.ly/3aBT2Ls
- Boonyaratpalin S, Supamattaya K, Kasornchandra J, Direkbusaracom S, Aekpanithanpong U, et al. (1993) Non-occluded baculo-like virus, the causative agent of yellow head disease in the black tiger shrimp (Penaeus monodon). Fish Pathol 28: 103-109. Link: https://bit.ly/324aeWd
- Spann KM, Vickers JE, Lester RJ (1995) Lymphoid organ virus of Penaeus monodon from Australia. Dis Aquat Org 23: 127-134. Link: https://bit.ly/3iXF4H0
- Zhang S, Bonami JR (2007) A roni‐like virus associated with mortalities of the freshwater crab, Eriocheir sinensis Milne Edwards, cultured in China, exhibiting ‘sighs disease’and black gill syndrome. J Fish Dis 30: 181-186. Link: https://bit.ly/31ajvg3
- Bossart GD, Schwartz JC (1990) Acute necrotizing enteritis associated with suspected Coronavirus infection in three harbor seals (Phoca vitulina). J Zoo Wildl Med 21: 84-87. Link: https://bit.ly/2CJNKRK
- Mihindukulasuriya KA, Wu G, Leger JS, Nordhausen RW, Wang D (2008) Identification of a novel Coronavirus from a beluga whale by using a panviral microarray. J Virol 82: 5084-5088. Link: https://bit.ly/3iV6ZHw
- Woo PC, Lau SK, Lam CS, Tsang AK, Hui SW, et al. (2014) Discovery of a novel bottlenose dolphin Coronavirus reveals a distinct species of marine mammal Coronavirus in GammaCoronavirus. J Virol 88: 1318-1331. Link: https://bit.ly/3heQfKE
- De Groot RJ, Cowley JA, Enjuanes L, Faaberg KS, Perlman S, et al. (2011) Order nidovirales. Virus taxonomy 2011.
- Cong Y, Verlhac P, Reggiori F (2017) The interaction between nidovirales and autophagy components. Viruses 9: 182. Link: https://bit.ly/2Yh3Jhz
- Sawicki SG, Sawicki DL, Siddell SG (2007) A contemporary view of Coronavirus transcription. J Virol 81: 20-29. Link: https://bit.ly/2E6w8jD
- Fichtner D, Granzow H, Dauber M, Schütze H, Bergmann S, et al. (2005) Further data on the “white bream virus”, a novel virus from fish: immunoelectron and immunofluorescence studies with monoclonal antibodies. X. Gemeninschaftstagungder Deutschen, der Österreichischen und der Schweizer Sektion der European Association of Fish Pathologists (EAFP) 2005.
- Fichtner D, Bergmann SM, Dauber M, Enzmann PJ, Granzow H, et al. (2009) Dituation of fish epidemics in Germany and selected case reports from the National Reference Laboratory. XII. Gemeninschaftstagung der Deutschen, der Österreichischen und der Schweizer Sektion der European Association of Fish Pathologists (EAFP).
- Faisal M, Baird A, Winters AD, Millard EV, Marcquenski S, et al. (2016) Isolation of the fathead minnow nidovirus from muskellunge experiencing lingering mortality. J Aquat Anim Health 28: 131-141. Link: https://bit.ly/323jTME
- McCann RL (2015) Viral survey of fathead minnows, golden shiners, and white suckers form baitfish dealers in Wisconsin (Doctoral dissertation). Link: https://bit.ly/2Q3fnrU
- Baird A, Faisal M (2016) Fathead minnow nidovirus infects spotfin shiner Cyprinella spiloptera and golden shiner Notemigonus crysoleucas. Dis Aquat Org 119: 37-44. Link: https://bit.ly/2EkVJVW
- Zhang Q, Standish I, Winters AD, Puzach C, Ulferts R, et al. (2014) Development and evaluation of reverse transcription loop-mediated isothermal amplification assay for the detection of the fathead minnow nidovirus. J Virol Methods 202: 39-45. Link: https://bit.ly/3hhZZEg
- Xiao-Yu C, Yong Z, Xin C, Jian Z, Xian-Dong Z, et al. (2019) Isolation and genetic analysis of a nidovirus from crucian carp (Carassius auratus). Arch Virol 164: 1651-1654. Link: https://bit.ly/3g8sHG1
- Mordecai GJ, Miller KM, Di Cicco E, Schulze AD, Kaukinen KH, et al. (2019) Endangered wild salmon infected by newly discovered viruses. Elife. Link: https://bit.ly/3aDRn86
- Limsuwan C (1991) Handbook for cultivation of black tiger prawns. Tansetakit, Bangkok, Thailand.
- Flegel TW (1997) Progress in research on yellow-head virus and white-spot virus in Thailand. Diseases in Asian aquaculture.
- OIE (World Organization for Animal Health) (2003). Manual of diagnostic tests for aquatic animals 2003.
- Flegel TW (1995) Environmental control of infectious diseases in Thailand. In Diseases in Asian aquaculture II: Proceedings of the Second Symposium on Diseases in Asian Aquaculture. Fish Health Section, Asian Fisheries Society.
- Lightner DV (1996) A handbook of shrimp pathology and diagnostic procedures for diseases of cultured penaeid shrimp. Link: https://bit.ly/3g876O9
- Spann KM, McCulloch RJ, Cowley JA, East IJ, Walker PJ (2003) Detection of gill-associated virus (GAV) by in situ hybridization during acute and chronic infections of Penaeus monodon and P. esculentus. Dis Aquat Org 56: 1-10. Link: https://bit.ly/2FEoSfi
- Walker PJ, Cowley JA, Spann KM, Hodgson RA, Hall MR, et al. (2001) Yellow head complex viruses: transmission cycles and topographical distribution in the Asia-Pacific region. Link: https://bit.ly/2Q2Z7Y2
- Cowley JA, Dimmock CM, Wongteerasupaya C, Boonsaeng V, Panyim S, et al. (1999) Yellow head virus from Thailand and gill-associated virus from Australia are closely related but distinct prawn viruses. Dis Aquat Org 36: 153-157. Link: https://bit.ly/2Q9kdUF
- Cowley JA (2016) Nidoviruses of Fish and Crustaceans. In Aquaculture Virology 443-472.Link: https://bit.ly/31cXRIh
- Bonami JR, Zhang S (2011) Viral diseases in commercially exploited crabs: a review. J Invertebr Pathol 106: 6-17. Link: https://bit.ly/3aE2vBV
- Duangsuwan P, Tinikul, Y, Withyachumnarnkul B, Chotwiwatthanakun C, Sobhon P (2011) Cellular targets and pathways of yellow head virus infection in lymphoid organ of Penaeus monodon as studied by transmission electron microscopy. Songklanakarin J Sci Technol 33: 2. Link: https://bit.ly/3iYuFe7
- Jahromi SS (1977) Occurrence of rhabdovirus-like particles in the blue crab, Callinectes sapidus. J Gen Virol 36: 485-493. Link: https://bit.ly/318YUZt
- Johnson PT (1983) Diseases caused by viruses, rickettsiae, bacteria, and fungi. The Biology of the Crustacea Pathobiology 6: 2-78.
- Yudin AI, Clark WH (1978) Two viruslike particles found in the ecdysial gland of the blue crab, Callinectes sapidus. J Invertebr Pathol 32: 219-221. Link: https://bit.ly/2Ea7QFg
- Yudin AI, Clark WH (1979) A description of rhabdovirus-like particles in the mandibular gland of the blue crab. Callinectes sapidus J Invertebr Pathol 33: 133-147. Link: https://bit.ly/2Q6gCGL
- Nollens HH, Wellehan JF, Archer L, Lowenstine LJ, Gulland FM (2010) Detection of a respiratory Coronavirus from tissues archived during a pneumonia epizootic in free-ranging Pacific harbor seals Phoca vitulina richardsii. Dis Aquat Org 90: 113-120. Link: https://bit.ly/3aFBuhO
- Zheng J (2020) SARS-CoV-2: an emerging Coronavirus that causes a global threat. Int J biol Sci 16: 1678-1685. Link: https://bit.ly/2Qan8MA
- Zhang T, Wu Q, Zhang Z (2020) Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr Biol 30: 1346-1351. Link: https://bit.ly/3l0pLz2
- Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, et al. (2003) Isolation and characterization of viruses related to the SARS Coronavirus from animals in southern China. Science 302: 276-278. Link: https://bit.ly/31cJL9Q
- Azhar EI, El-Kafrawy SA, Farraj SA, Hassan AM, Al-Saeed MS, et al. (2016) Evidence for camel-to-human transmission of MERS Coronavirus. N Engl J Med 370: 2499-2505. Link: https://bit.ly/3aE73s1
- Cui J, Li F, Shi ZL (2019) Origin and evolution of pathogenic Coronaviruses. Nat Rev Microbiol 17:181-192. Link: https://go.nature.com/3hcSPki
- Bondad-Reantaso MG, Mackinnon B, Bin H, Jie H, Tang-Nelson K, et al. (2020) SARS-CoV-2 (The Cause of COVID-19 in Humans) is Not Known to Infect Aquatic Food Animals nor Contaminate Their Products. Asian Fish Sci 33. Link: https://bit.ly/34eX9vK
- Consumption of fish, fishery products safe: Department of Fisheries on Coronavirus. Link: https://bit.ly/3aHNlfg
- Fehr AR, Perlman S (2015) Coronaviruses: an overview of their replication and pathogenesis. In Coronaviruses Humana Press. 1282: 1-23. Link: https://bit.ly/31aqHZJ
- Enjuanes L, Gorbalenya AE, De Groot RJ, Cowley JA, et al. (2008) Nidovirales. Encyclopedia of Virology 419-430. Link: https://bit.ly/31aPe0O
- OIE (World Organization for Animal Health) (2020) Infection with SARS-COV-2 in animals. Link: https://bit.ly/2EjvOhi

Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
