International Journal of Aquaculture and Fishery Sciences
Comparison of the Crossbreeding Effects of three Mandarin Fish populations and analyses of the Microsatellite Loci Associated with the growth traits of F1 Progenies
Qingkai Zeng1,2, Chengfei Sun1, Junjian Dong1, Yuanyuan Tian1 and Xing Ye1*
2College of Fisheries and Life Science, Shanghai Ocean University, Shanghai 201306, China
Cite this as
Zeng Q, Sun C, Dong J, Tian Y, Ye X (2017) Comparison of the Crossbreeding Effects of Three Mandarin Fish Populations and Analyses of the Microsatellite Loci Associated with the Growth Traits of F1 Progenies. Int J Aquac Fish Sci 3(2): 035-041. DOI: 10.17352/2455-8400.000026Cross breeding with different populations might lead to heterosis and enhance the genetic diversity of the resulting offspring. In this study, three populations of mandarin fish (Siniperca chuatsi), including two cultured (A and B) and one wild population (C), were used to construct three pure groups (A♂×A♀, B♂×B♀, C♂×C♀) and six crossbred groups (A♂×B♀, A♂×C♀, B♂×A♀, B♂×C♀, C♂×A♀, C♂×B♀). Growth performance was compared among all combinations, and 11 microsatellites were used to analyze the genetic diversity of the resulting offspring and identify correlations associated with growth traits (body weight, total length, and body height). The best growth performance values associated with the growth traits were detected in the progeny of the inbred A♂×A♀ group. The A♂×B♀ and A♂×C♀ F1 exhibited relatively high increases of growth in all crossbreed combinations, whereas the F1 of C♂×B♀ exhibited lower growth performance. Cross combinations that used male fish from the A population displayed significant growth advantages. Analysis of genetic diversity showed that the expected heterozygosity (He) of the F1 of all combinations were greater than 0.5 except A♂×C♀, B♂×A♀, and A♂×A♀. The results of association analyses indicated that seven microsatellite loci, est21, w19521, sch14, sc90, est43, w19517, and sc01 were associated with the targeted growth traits. This result laid a good foundation for further breeding selection of mandarin fish using population selection and molecular marker-assisted selection. It also would provide more reliable guidelines for breeding selection of other fish species.
Introduction
Crossbreeding refers to the method based on genetic recombination, genetic separation, and progeny selection that is used to cultivate new breeds with concentrated favorable genes via crossbreeding. Crossbreeding not only leads to heterosis but also enriches the genetic structure of a population [1]. The cultivation of common carp (Cyprinus carpio) is the most successful example of the crossbreeding method. For instance, carp cultivated in the Soviet Union (e.g. Ropsha carp, Ukrainian-Ropsha carp, and Krasnodar carp) [2] and those cultivated in China (e.g. Feng carp, Yue-carp, and Heyuan carp) [3-5], were successfully bred via crossbreeding. Therefore, these examples provide evidence that crossbreeding is an effective method for breeding aquatic organisms. Regarding other cultivated fishes [6], the crossbreeding of two wild strains of silver perch from Cataract Dam (C) and Murray River (M) indicated that M♂×C♀ was 20.9% and 16.0% heavier than the mid-parent and best-parent averages, respectively, and prominent heterosis was detected [6]. Furthermore, crossbreeding of the Donaldson strain of rainbow trout (Oncorhynchus mykiss) exhibited the most positive growth performance, deduced based on the results of a complete diallel cross with Bohai, Denmark, Donaldson, Norway, and California strains [7]. Crossbreeding and the resulting heterosis have also been examined in other cultured species. For example, a complete diallel cross between India (II0) and Sanya (SS0) populations of pearl oyster (Pinctada martensii) resulted in the construction of four groups of offspring, and the offspring of II0♀×SS0♂ and SS0♀×II0♂ exhibited heterosis in shell length, shell width, shell height, hinge length, and shell weight [8]. Recently, several new crossbred breeds were certified by the National Certification Committee for Aquatic Varieties of China, including “New Gift” Nile tilapia (Oreochromis niloticus) [9], “ping you No.1” Paralichthys olivaceus [10] and “Turbot No.1” Scophthalmus maximus [11].
Marker-assisted selection is an indirect selection process where a trait of interest is selected based on a specific marker that is linked to that trait. The practice of selecting traits of interests at the molecular level negates environmental effects and results in reliable data [12]. For example, the utilization of sex-linked markers led to the successful cultivation of “All-male No.1” yellow catfish (Pelteobagrus fulvidraco), all-female “Northern Flounder No.1” and “Northern Flounder No.2” flounder, “Luxiong 1” Nile tilapia, and the high-female fry of half-smooth tongue sole (Cynoglossus semilaevis) [13]. Microsatellites exhibit high polymorphism rates, high abundance, and a broad distribution throughout the genome, and they represent one of the most popular genetic markers for genetic diversity evaluation, parentage analysis, and molecular marker-assisted breeding [14]. In previous studies on genetic breeding of aquatic organisms, correlations between microsatellites and growth traits have been detected. For instance, eight microsatellite markers were obtained from 65 Nile tilapia microsatellites using correlation analyses of growth traits, and three of these can be reliably used for molecular marker-assisted selection of Nile tilapia [15]. Twelve and 13 microsatellites of crucian carp (Carassius auratus) were significantly correlated with body weight and body length, respectively, and eight of these were significantly correlated with both body weight and length [16]. Furthermore, 29 microsatellites were selected from pearl oyster expressed sequence tags (ESTs) for correlation analyses, and three were found to be significantly correlated with phenotypic traits [17]. Microsatellites that were correlated with phenotypic traits were also detected in common carp [18], large yellow croaker (Larimichthys crocea) [19] and mud crab (Scylla serrata) [20] populations.
Mandarin fish (Siniperca chuatsi) is one of the most important cultured freshwater fish in China, and the lack of selective breeding of this species in recent decades has resulted in a decline in the growth rate of pond-cultured fish. In addition, shortened period of sexual maturity and reduced anti-disease ability have seriously affected the quality and safety of mandarin fish aquaculture. Therefore, it is necessary to establish a selective breeding program for mandarin fish. In the present study, three mandarin fish populations, including two cultured populations (Guangdong [A] and Anhui [B]) and one wild population (Hunan [C]), were used to construct three pure groups and six crossbred groups. Growth performance and genetic diversity were compared among all the combinations, and correlations between microsatellite markers and growth traits were determined in order to provide a solid foundation for the molecular-assisted selection of mandarin fish.
Materials and Methods
Ethics statement
All the handling of fishes was conducted in accordance with the guidelines on the care and use of animals for scientific purposes set up by Institutional Animal Care and Use Committee (IACUC) of the Pearl River Fisheries Institute, Chinese Academy of Fishery Sciences. The IACUC specially approved this study under the project “Breeding of mandarin fish”.
Materials
Cultured Guangdong (A) and Anhui (B) mandarin fish populations, introduced in 2004 and 2013, respectively, were provided by the Qingyuan Yushun Farming and Fishery Science and Technology Service Limited Corporation. The Hunan (C) wild population was collected from Dongting Lake, Hunan.
Experimental methods
Mating design and family production: Breeding experiments were conducted at the breeding base of the Qingyuan Yushun Farming and Fishery Science and Technology Service Limited Corporation. Each parent was injected with oxytocin (Ningbo Sansheng Pharmaceutical Co., Ltd., China) to artificially induce spawning. Three pure groups (A♂×A♀, C♂×C♀, B♂×B♀) and six crossbred groups (A♂×B♀, A♂×C♀, B♂×A♀, B♂×C♀, C♂×A♀, C♂×B♀) were constructed in this study (Table 1). All parents were divided among the nine hatchery ponds (4π × 1 m3) to spawn and hatch, according to the pairing mode listed in table 1. The fry of the nine groups were subsequently transferred to nine cement pools (3 × 2 × 1 m3) with uniform flow, continuous oxygenation, and small living fish fry for cultivation at approximately 28 °C.
Approximately 350 fry were randomly selected from each group when their total length reached approximately 12 cm. Microchips (ID100A, Trovan, UK) were used to mark the fry, and an ARE H5 hand-held reader was used to scan and record the encoded data. We then measured and recorded the body weight (BW), body height (BH), and total length (TL) of each fish. All marked fry were cultivated in a pool (1000 m2 × 1.5 m), and all marked specimens were recaptured following a four-month cultivation period. Microchips were then scanned, and BW, BH, and TL were measured and analysed to assess the growth statuses of all groups. Approximately 80–90 specimens from each group were selected, and their fin samples were obtained and preserved in 95% ethanol prior to DNA extraction.
Polymerase chain reaction (PCR) amplification and detection of the amplified PCR products.
Genomic DNA was extracted using a TIANamp Marine Animal DNA Kit (Tiangen Biotech CO., LTD., Beijing) from the fin samples (approximately 30 mg). The fin sample was digested in a tube with 200 μL buffer GA and 20 μL proteinase K for 3 h at 56 °C. Then 200 μl GB and 200 μl ethanol were added in successively, fully invert and mix evenly. The mixed liquid was moved into an adsorption column, then centrifuged. The column was rinsed by buffer GD and buffer PW, and then centrifuged. The DNA was finally eluted by 80 μL ddH2O. DNA quality (1.8 < OD260/OD280< 2.0) was assessed using electrophoresis on a 1% agarose gel and a spectrometer. Extracted DNA was preserved in a refrigerator at −20 °C until further analyses.
Thirty primer pairs for mandarin fish microsatellite sequences were designed based on sequences found in GenBank, and each primer pair was synthesized at Guangzhou Ige Biotechnology LTD. Twelve random samples were used to detect the specificity and polymorphism of each primer set, and 11 primer pairs were confirmed for this study (Table 2).
The total PCR amplification volume was 20 μL, including 10 μL of TaKaRa Premix Taq (TaKaRa, Dalian, China), 1 μL each of the forward and reverse primers, 1 μL of DNA template, and 7 μL of ddH2O. PCR was performed in 96-well plates using the following protocol: denaturation for 5 min at 94 °C; 32 cycles of 94 °C for 30 s, annealing (temperatures indicated in table 2) for 30 s, 72 °C for 30 s; and a final extension at 72 °C for 10 min. The amplified products were stored at 4 °C.
The amplified PCR products were centrifuged for 1 min at 2000 g, and were then incubated for 2–3 min at room temperature. The products were carefully inspected to ensure that bubbles were not present, and 10 μL of mineral oil was then used to cover each product to prevent evaporation of the sample. The sample was then placed in a Qsep100 analyzer, and the following electrophoretic parameters were set: sample injection at 6 kV for 2 s and electrophoresis at 6 kV for 300 s. A Q-Analyzer was used to analyze and adjust the results of the electrophoretic analysis, and genotyping of microsatellite loci was performed for each individual based on the differences in the molecular weights of the amplified products.
Statistics and analysis
The number of alleles (Na), number of effective alleles (Ne), Shannon index (I), observed heterozygosity (Ho), and expected heterozygosity (He) of each population were determined using GenAlex 6.502, and CERVUS 3.0 was used to analysis the polymorphic information content (PIC).
The mean and standard deviation values of BW, BH, and TL were calculated using SPSS 21.0, and the data were used to compare growth statuses. The equation used to determine the heterosis rate (H) was as follows:
F1 Represents the mean growth traits of the crossbreed population, P1 and P2 represent the mean growth traits of the respective purebred populations [21].
One-hundred and twenty samples were randomly selected for growth association analyses of microsatellite markers. To examine the relationship between genotypes and growth traits, a general linear model was used to conduct multiple comparisons of genotypes and specific alleles, and each genotype was represented by more than three samples.
Results
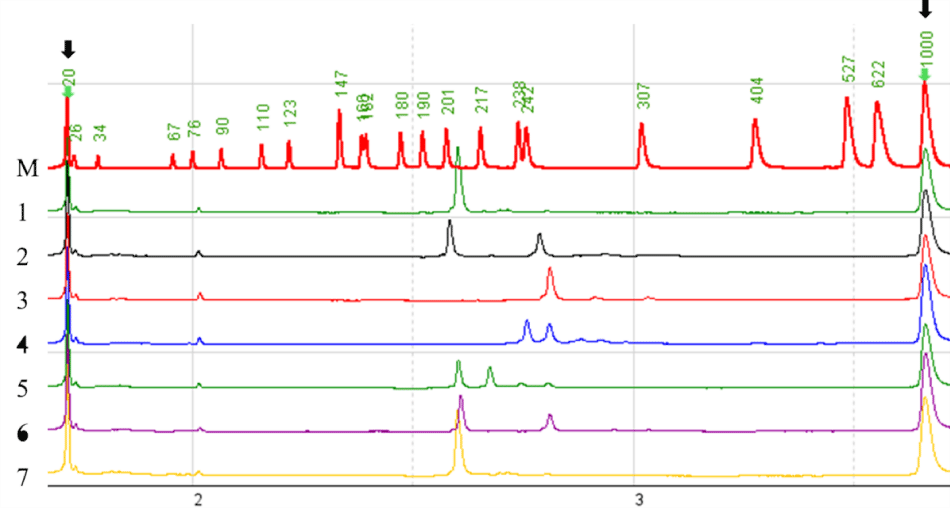
Detection of PCR amplification products via capillary electrophoresis
PCR amplification products were detected via capillary electrophoresis (Figure 1). Regarding the 11 microsatellite loci, significant fluorescence signals and polymorphisms were detected, and 75 alleles are obtained for all loci.
Genetic diversity of each F1 generation
The genetic diversity parameters of each F1 generation are shown in table 3. With the exception of the F1 generations of A♂×C♀, B♂×A♀, and A♂×A♀, the He values of all F1 progeny were greater than 0.5. The F1 progeny of C♂×B♀ exhibited the highest genetic diversity (>He = 0.543), while the F1 progeny of A♂×A♀ displayed the lowest genetic diversity (>He = 0.462).
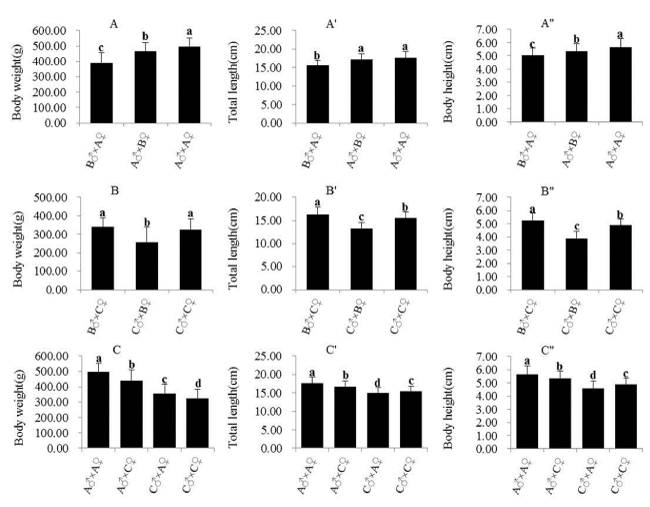
Comparisons of growth among each population
The growth performances among the inbred and crossbred populations are showed in figure 2. The growth traits (BW, BH, and TL) of A♂×A♂ exhibited the best performance among all populations. Furthermore, A♂×B♀ and A♂×C♀ performed better among the crossbred populations, while C♂×B♀ performed the worst. In addition, the heterosis rates of BW, TL, and BH were 6.64%, 6.64%, and 1.33%, respectively, in A♂×C♀ and −33.20%, −21.55%, and −27.81% in C♂×A♀, respectively (Table 4).
Association analysis of growth traits
Table 5 shows the normal distribution of the BW, TL, and BH test results for 120 samples. The three growth traits of mandarin fish corresponded to the normal distribution based on the Kolmogorov-Smirnov single sample normal distribution test (P > 0.05).
Genotypes with significant effects of growth traits were found in seven microsatellites (Table 6). BC of w19521 was the dominant genotype associated with body weight. AB of est21, BC of w19521, DD of sch14, BC of sc90, and BC of est43 were dominant genotypes associated with total length. Lastly, AB of est21, AB of w19517, BC of w19521, DD and BD of sch14, and AB of sc01 were dominant genotypes associated with body height.
Discussion
Comparison of the crossbreeding effects of the three mandarin fish populations
Heterosis refers to the phenomenon in which the progeny of diverse varieties of a species or crosses between species exhibit greater biomass, speed of development, and fertility than the progeny of the parental varieties [22]. In the present study, the growth traits (BW, BH, and TL) of A♂×A♀ exhibited the best performance among all the populations. Moreover, A♂×B♀ and A♂×C♀ performed better among the crossbred populations, indicating that the A population was the most suitable male parent for breeding. Furthermore, A♂×C♀ exhibited heterosis, while outbreeding depression was detected in C♂×A♀. This phenomenon was found in another crossbreeding study of mandarin fish, which examined wild Hunan (HC) and Jiangsu (JC) crossbred populations, and the growth traits indicated obvious outbreeding depression (JC×JC > HC×HC > HC×JC > JC×HC) [23]. Outbreeding depression has also been observed in other crossbred fish, including Chinook salmon (Oncorhynchus tshawytscha) [24] and brook trout (Salvelinus fontinalis) [25].
In the present study, the genetic diversity of the C♂×B♀ and A♂×A♀ populations were the highest and lowest among all populations, respectively. Furthermore, the progeny of a male or female parents exhibited relatively low genetic diversity, but the values were higher than those observed in inbred populations. However, it was widely reported that an extended period of breeding and intensive selection might cause allelic loss, which could reduce genetic diversity [26]. Species with abundant genetic diversity and variability adapt easily to variable environments, and genetic diversity is also closely associated with the vitality and breeding of a species. Therefore, species with higher genetic diversity have greater evolutionary potential [27]. In the present study, the A population experienced more years of breeding and selection compared with the two other populations, according to a previous analysis, the genetic diversity of these three groups decreased in the following order: C > B > A [28]. In the present study, the pure strain of A population exhibited the greatest growth performance, but its genetic diversity was the lowest. Through crossbreeding, genetic diversity was improved, which in turn improved the breeding potential, thereby laying the foundation for the sustainable development of mandarin fish.
Correlation analysis between the growth traits and microsatellite loci of mandarin fish F1 progenySome of the important economic traits of fish (e.g. growth and disease resistance) are usually controlled by multiple genes [29]. In common carp, microsatellite loci HLJ133, HLJ346, and HLJ360 were found to be significantly correlated with BW, and HLJ133, HLJ368, and HLJ390 were found to be correlated with BL [18]. In crucian carp, 12 microsatellite loci (e.g. JE134, JE180, and JE253) simultaneously controlled BW, and 13 microsatellite loci (e.g. JE20, JE134, JE588, and JE1108) were significantly associated with BL [16]. In the present study, est21, w19521, sch14, sc01, sc90, and est43 were significantly correlated with TL, and BH was controlled by est21, w19517, w19521, sch14, and sc01. Additionally, one locus could control several growth traits via pleiotropic effects. For example, w19521 was correlated with BW, TL, and BH, but the close association between BW, TL, and BH might cause this phenomenon. A similar phenomenon was also detected in other fish species, where UNH914 and UNH974 were found to be significantly associated with the BW, BL, and BH of Nile tilapia [15] and JE134, JE588, and JE1108 that were correlated with the body weight and body length of crucian carp [16].
Quantitative traits are generally controlled by multiple genes, but only a few “major genes” have powerful effects [30-32]. In the present study, seven microsatellite loci that were correlated with growth traits were selected from 11 microsatellite loci, including one, five, and six genotypes with positive effects on BW, TL, and BH, respectively. To provide more accurate and effective guidance for molecular marker-assisted breeding of mandarin fish, these seven screened loci will be further verified to confirm their role as “major loci” in future studies.
In conclusion, combinations of good growth performances were obtained via the crossbreeding of three mandarin fish populations, and the genetic diversity of the breeding populations were also enriched via crossbreeding. Microsatellite loci associated with growth traits and genotypes with positive effects on those growth traits were identified using correlation analyses. Thus, the data obtained in the present study laid a good foundation for further breeding selection of mandarin fish using population selection and molecular marker-assisted selection. It also would provide more reliable guidelines for breeding selection of other fish species.
This work was supported by grants from the Science and Technology Project of Guangdong province (No. 2015A020209034), Ocean Fisheries Science and Technology Promotion Project of Guangdong province (No. A201601A06), Science and Technology Project of Guangzhou (No. 2014J4100087), China Agriculture Research System (CARS-49).
- Lou YD (2006) Fish Breeding. In: Cross Breeding (ed. by J.B. Shen & M.H. Liu). China Agriculture Press, Beijing, pp. 40-106.
- Kirpichnikov VS, Ilyasov JI, Shart LA, Vikhman AA, Ganchenko MV, et al. (1993) Selection of krasnodar common carp (Cyprinus carpio L.) for resistance to dropsy: principal results and prospects. Aquaculture 111: 7-20. Link: https://goo.gl/fBhpkt
- Ma Z, Zhang X, Qiu Q, Liu S, Zhang J (1981) The analysis of the economical characteristics of a hybrid from two ecological types of carps. Journal of Fisheries of China 29: 38-45. Link: https://goo.gl/c9JCVq
- Li W (1988) Study on LDH isoenzymes of hybrid yue-carp and its parents. Acta Genetica Sinica 15: 46-51. Link: https://goo.gl/52Q8za
- Yang YO, Xie SQ, Yao F (2005) Activity levels and rhythms of Feng carp (Cyprinus carpio var. xingguo♀×Cyprinus carpio var. mirror splittered♂) and its parents on starvation and satiation. Acta Zoologica Sinica 51: 806-812. Link: https://goo.gl/PefDa2
- Guy JA, Jerry DR, Rowland SJ (2009) Heterosis in fingerlings from a diallel cross between two wild strains of silver perch (Bidyanus bidyanus). Aquaculture Research 40: 1291–1300. Link: https://goo.gl/DajbUv
- Hu G, Gu W, Yang R, Wang B (2016) Multivariate diallel cross-analyses for body weight of rainbow trout, Oncorhynchus mykiss, in China. Journal of the World Aquaculture Society. Link: https://goo.gl/PUQJif
- Wang AM, Wang Y, Gu ZF, Li M, Shi YH, et al. (2010) Heterosis and genetic variation of hybrids from two geographical populations of pearl oyster, Pinctada martensii. Dongwuxue Yanjiu 41: 140-147. Link: https://goo.gl/bHuKQU
- Li SF, Tang SJ, Cai WQ (2010) RAPD-SCAR markers for genetically improved new gift nile tilapia (Oreochromis niloticus niloticus l.) and their application in strain identification. Dongwuxue Yanjiu 31: 147-153. Link: https://goo.gl/EmE5Ff
- Liu F, Chen S, Wang L, Tian Y, Deng H, et al. (2013) Analysis of growth performance and breeding value of “ping you no.1” Japanese flounder and selection of parents. Journal of Fishery Sciences of China 20: 521-527. Link: https://goo.gl/7VueTg
- Ma AJ, Wang XA, Yang Z, Qu JB, Lei QL, et al. (2016) “Duobao No. 1” Turbot. China Fisheries 61-63.
- Francia E, Tacconi G, Crosatti C, Barabaschi D, Bulgarelli D, et al. (2005) Marker assisted selection in crop plants. Plant Cell Tissue & Organ Culture 82: 317-342. Link: https://goo.gl/uUx1HF
- Gui JF, Bao ZM, Zhang XJ (2016) Development strategy for aquaculture genetic breeding and seed industry. Engineering Science 18: 8-14. Link: https://goo.gl/YWTxne
- Miah G, Rafii M, Ismail M, Puteh A, Rahim H, et al. (2013) a review of microsatellite markers and their applications in rice breeding programs to improve blast disease resistance. Int J Mol Sci 14: 22499-22528. Link: https://goo.gl/FFnfi3
- Liu FP, Bai JJ, Song HM, Ye X, Li SJ, et al. (2010) Correlation analysis of microsatellite DNA markers with major growth traits of tilapia (Oreochromis niloticus). Journal of Fisheries of China 34: 169-177. Link: https://goo.gl/4ciLhC
- Zheng X, Kuang Y, Lü W, Cao D, Sun X (2014) Transcriptome-derived EST–SSR markers and their correlations with growth traits in crucian carp Carassius auratus. Fisheries Science 80: 977-984. Link: https://goo.gl/g2WFTt
- Qiu Y, Lu H, Zhu J, Chen X, Wang A, et al. (2014) Characterization of novel EST-SSR markers and their correlations with growth and nacreous secretion traits in the pearl oyster Pinctada martensii (dunker). Aquaculture 420-421: S92–S97. Link: https://goo.gl/X7Q72T
- Chu ZY, Zhang XF, Cao Z, Sun XW (2011) Correlation analysis of the 4 growth traits in the common carp backcross population. Journal of Fisheries of China 35: 10-19. Link: https://goo.gl/Z5aVgR
- Liu F, Yao J, Wang X, Repnikova A, Galanin DA, et al. (2012) Genetic diversity and structure within and between wild and cultivated Saccharina japonica, (Laminariales, Phaeophyta) revealed by SSR markers. Aquaculture S358–359: 139-145. Link: https://goo.gl/Rs8GFU
- Ma H, Jiang W, Liu P, Feng N, Ma Q, et al. (2014) Identification of transcriptome-derived microsatellite markers and their association with the growth performance of the mud crab (Scylla paramamosain). PLoS One 9: e89134. Link: https://goo.gl/dvtxxQ
- Tave D (1993) Genetics for fish hatchery managers (ed.). Van Nodtrand Reinhold 212-213. Link: https://goo.gl/K6rXqS
- Li WT (2000) Animal Genetics and Breeding. In: Utilization of Heterosis. China Agricultural University press p: 265-284.
- Sun JJ, He JG, Liu L, Wang HF, Lu X, et al. (2016) Analysis of growth and genetic characteristics of the breeding populations of Siniperca chuatsi. Journal of Fishery Sciences of China 23: 425-435. Link: https://goo.gl/Akv9DG
- Bryden CA, Heath JW, Heath DD (2004) Performance and heterosis in farmed and wild Chinook salmon (Oncorhynchus tshawytscha) hybrid and purebred crosses. Aquaculture 235: 249-261. Link: https://goo.gl/nfP7Gy
- Granier SG, Audet CA, Bernatchez LB (2010) Heterosis and outbreeding depression between strains of young-of-the-year brook trout (Salvelinus fontinalis). Canadian Journal of Zoology 89: 190-198. Link: https://goo.gl/qrPeYH
- Liu XD, Wei XJ, Cai MY, Liu Y, Wang ZY (2012) The segregation patterns of 22 microsatellite markers in an F1 family of large yellow croaker (Larimichthys crocea) and correlation analysis with growth-related traits. Journal of Fisheries of China 36: 1322-1330.
- Ji WZ, Su B (1999) Principles and Methodologies of Genetic Diversity Studies. In: Summary (ed. by L. M. Shi, B. Su & Y. P. Zhang). Zhejiang Science and Technology Publishing House, Hangzhou p: 1-12.
- Zeng QK, Sun CF, Dong JJ, Tian YY, Shi HY, Jiao ZZ, Ye X (2017) Analysis of genetic diversity in three different populations of Siniperca chuatsi. Genomics and Applied Biology, in press.
- Tong JG, Sun XW (2015) Genetic and genomic analyses for economically important traits and their applications in molecular breeding of cultured fish. Sci China Life Sci 58: 178-186. Link: https://goo.gl/fnXUQq
- Hedgecock D, Davis JP (2007) Heterosis for yield and crossbreeding of the Pacific oyster Crassostrea gigas. Aquaculture 272: S17-S29. Link: https://goo.gl/A8x4TN
- Thanh NM, Nguyen NH, Ponzoni RW, Vu NT, Barnes AC, et al. (2010) Estimates of strain additive and non-additive genetic effects for growth traits in a diallel cross of three strains of giant freshwater prawn (Macrobrachium rosenbergii) in Vietnam. Aquaculture 299: 30-36. Link: https://goo.gl/u8uKpC
- Wang C, Li Z (2010) Improvement in production traits by mass spawning type crossbreeding in bay scallops. Aquaculture 299: 51-56. Link: https://goo.gl/N1BeXn

Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.
 Save to Mendeley
Save to Mendeley
