Global Journal of Perioperative Medicine
Machine learning characterization of a novel panel for metastatic prediction in breast cancer
Melih Ağraz1*, Umut Ağyüz2, E Celeste Welch3, Birol Kuyumcu4, and M Furkan Burak5,6
2Genz Biotechnology, 5699 Sk No:7/4 Çankaya, Ankara, Turkey
3Brown University, Center for Biomedical Engineering, 184 Hope Street, Providence, USA
4Sefa Merve RD Center, Cevizli, Akasya Sk. No:67, 34846 Maltepe, Istanbul, Türkiye
5Division of Endocrinology, Diabetes, and Hypertension, Brigham & Women’s Hospital and Harvard Medical School, 221 Longwood Avenue, Boston, MA 02115, USA
6Department of Molecular Metabolism, Harvard T.H. Chan School of Public Health, Boston, MA 02115, USA
Cite this as
Ağraz M, Ağyüz U, Welch EC, Kuyumcu B, Burak MF (2022) Machine learning characterization of a novel panel for metastatic prediction in breast cancer. Glob J Perioperative Med 6(1): 005-011. DOI: 10.17352/gjpm.000011Copyright License
© 2022 Ağraz M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Metastasis is one of the most challenging problems in cancer diagnosis and treatment, as causal factors have yet to be fully disentangled. Prediction of the metastatic status of breast cancer is important for informing treatment protocols and reducing mortality. However, the systems biology behind metastasis is complex and driven by a variety of interacting factors. Furthermore, the prediction of cancer metastasis is a challenging task due to the variation in parameters and conditions specific to individual patients and mutation subtypes.
In this paper, we apply tree-based machine learning algorithms for gene expression data analysis in the estimation of metastatic potentials within a group of 490 breast cancer patients. Tree-based machine learning algorithms including decision trees, gradient boosting, and extremely randomized trees are used to assess the variable importance of different genes in breast cancer metastasis.
Highly accurate values were obtained from all three algorithms, with the gradient boosting method having the highest accuracy at 0.8901. The most significant ten genetic variables and fifteen gene functions in metastatic progression were identified. Respective importance scores and biological functions were also cataloged. Key genes in metastatic breast cancer progression include but are not limited to CD8, PB1, and THP-1.
Introduction
Metastasis begins with the displacement of tumor cells from the primary tumor. Circulating tumor cells (CTCs) move through the vascular system to a distant organ. There, they colonize the new environment, forming a new tumor.
Metastasis is one of the most complex and challenging problems in the cancer field as its main causes are multifaceted and not well-understood yet. Additionally, it is strongly correlated with mortality, making it the most critical area in need of research within the field of cancer diagnostics [1].
Metastasis begins with the loss of cell-to-cell and cell-to-matrix adhesion. This facilitates local infiltration of tumor cells into adjacent tissues as well as trans endothelial migration into vessels via the process of intravasation. Cancer cells must transform themselves from endothelial cells into mesenchymal cells, known as epithelial to mesenchymal transition (EMT). This process is characterized by the loss of cellular adhesive properties and polarity with a simultaneous gain of other properties that enable CTCs to migrate to distant organs, extravasate, proliferate and colonize a discrete competent organ. The other major factors for metastasis are cell adhesion defects, angiogenesis, and disrupted cell signaling and metabolism.
Disrupted cell signaling interrupts foreign recognition responses, allowing cancer cells to pass through the circulation without being recognized by the immune system. However, CTCs can evade immune recognition by mimicking peripheral immune tolerance, as recently detailed by Gonzalez et al. [2].
CTCs have abnormal gene expression characteristics that are different from the primary tumor and help improve their survival in circulation [3]. Survivin is a major member of the inhibitor of apoptosis family (IAP) and it facilitates the escape of tumor cells from immune recognition by blocking the cytotoxicity of NK cells and PD-L1. It can mediate the regulatory T-cells (Tregs) to play a role in immunosuppression.
Metastasis has most frequently been investigated in late-stage metastatic tumors, the products of colonization of discrete regions. It is still ambiguous how metastatic mechanisms begin in the primary tumor in the early stages and how an expression changes over time [4]. This is important not only from the basic science perspective but from the diagnostic and predictive perspective as well. Cancer mortality can be reduced when appropriate anti-metastatic treatments are started earlier. Until then, the inability to reliably characterize metastasis continues to drive cancer’s reputation as the most unpredictable and challenging illness to treat, resulting in lower-than-predicted survival times [5].
Machine learning is a combination of statistics and computer science which has become popular in recent years due to increases in computational power, data availability, and data quantity. Machine learning approaches have been used in different fields of bioscience such as in biological network representation [6], classification and diagnosis [7], medical status prediction [8] and more [9]. This approach has recently become popular specifically in bioinformatics and cancer research [10].
As machine learning capabilities grow, predictive models have become more and more accurate at determining cancer metastasis. For example, Huang, et al. [11] used support vector machine (SVM) and SVM Ensembles to predict breast cancer, Behravan, et al. [12] predicted breast cancer risk using machine learning algorithms for genetic and demographic datasets, Xiaoa, et al. [13], used deep learning in cancer prediction Kadir and Gleeson [14] implemented machine learning methods in the classification of lung cancer in images and Azzawi [15] conducted lung cancer prediction from microarray data. Decision trees are some of the most popular non-parametric supervised classification machine learning algorithms. They are used to classify the data in the form of an inverted tree that consists of a leaf node, root node, and internal node [16]. The extremely randomized trees model is a tree-based ensemble model which was first introduced by Geurts, et al. [17] 2006. This algorithm is similar to the random forest model which selects the subset of K features when deciding to split at each node. However, the difference between the random forest and extremely randomized trees (ERT) model is that ERT creates the trees from the learning samples. The Gradient Boosting tree model is an ensemble model technique thought to originate from the work of Breiman [18], which was later progressed by Friedman [19].
Due to the success of this approach in predicting and classifying different forms of biological data, we have opted to apply this method herein to analyze the metastatic gene expression data from breast cancer patients using large, publicly available datasets. The dataset contains information on the expression of 23397 genes across 490 individuals. The full datasets also contain significant amounts of other information, including cancer type, tumor grade, and age. t-statistics and the Bayesian method were first applied to select important predictors. The differential expression of genes between 2 groups: metastatic and non-metastatic were subsequently analyzed and profiled. A Differential Gene Expression (DGE) Analysis was performed between these 2 groups using R software. Using this analysis, 133 significant transcripts were detected with a >1.5-fold change. Significant dimensionality reduction was applied to simplify and better interpret the data. In the subsequent framework, the metastatic and non-metastatic expression profiles are further investigated using the previously mentioned machine learning models to determine significant metastatic predictors. Tree-based machine learning algorithms were first applied to the reduced candidate data following DGE. Variable importance was used to examine variable responses and thereby identify the variables that most influence breast cancer metastasis.
Materials and methods
There are two main aims of this study. The first one is to show which of the tree-based algorithms is the most efficient in array analysis, and the second is to demonstrate which transcript outputs of these algorithms are the most significant both biologically and for future modeling approaches.
To address the first aim, data were processed by various machine learning methods to assess which method possesses the highest accuracy for this type of analysis. Decision trees, gradient boosting and extremely randomized trees were tested and compared. Each model was able to report separate variables with the highest value of metastatic predictive capability.
2 different cohort studies were merged to create the single dataset that was used in this study. Publicly available datasets GSE102484 and GSE20685 were downloaded from NCBI GEO Databank (https://www.ncbi.nlm.nih.gov/geo/). Both datasets were obtained from the same microarray chip platform GPL570 [HG-U133 Plus2] Affymetrix Human Genome U133 Plus 2.0 Array chip platform. ll cancer patients were diagnosed with breast cancer of clinical stages I-III. The data originates from a cohort study of invasive breast carcinoma patients who underwent surgery. Genomic data were obtained by whole RNA study from fresh frozen samples stored at a cancer center in Taiwan. These samples were obtained from total mastectomy and sentinel lymph node biopsy procedures. Any patient pretreated by chemotherapy or radiotherapy was excluded from this cohort data. (n=683).
A second dataset GSE20685 was merged with the first. In this cohort study, genomic profiles were assessed from the whole RNA of fresh frozen samples obtained from patients diagnosed and treated with breast cancer between 1991–2004. The samples were stored at the National Cancer Center Singapore. Centroid analysis was used to determine molecular subtypes of breast cancer (n=312) [20].
These two datasets were combined in R. Mutual parameters and data points were selected for data alignment before the merge. 44 transcripts with missing (NA) values in more than 30% of observations were excluded from the data. Novel R programming codes for data manipulation and normalization were utilized instead of relying on built-in functions.
The Bioconductor RMA package and quantile normalization functions were applied for inter-array normalizations. After the combination, a consensus of 54643 transcripts was merged and preprocessed.
80% of the data was used to train the machine learning model while 20% was used to test. five-fold cross-validation was applied, and the model was then trained with the decision tree, extremely randomized tree, and gradient boosting approaches. These machine learning approaches were selected as tree structures are powerful in modeling and these particular approaches are able to represent the variable importance of genes within the model of the tree structures.
Then the precision, recall, F1-score, and accuracy were calculated for each model as accuracy measures. All relevant equations, such as the formula of accuracy measures can be seen in the supplementary document.
Results and discussion
The gradient boosting approach is the algorithm that has the most accurate results when compared to all others tested across a variety of metrics including precision, recall, F1 score, and accuracy (Table 1). In the first table, it can be seen that gradient boosting is able to predict whether a patient has metastatic cancer from the input expression data since it has the highest accuracy results for precision, recall, F1-score, and accuracy. The precision, recall, F1-score, and accuracy were calculated as 0.8901, 0.8550, 0.8666, and 0.8780 for this model, respectively.
Machine learning tree models can be used to determine the variables with the most predictive importance, helping algorithms to assign greater weight to data that plays more of a role in the proper classification of metastasis.
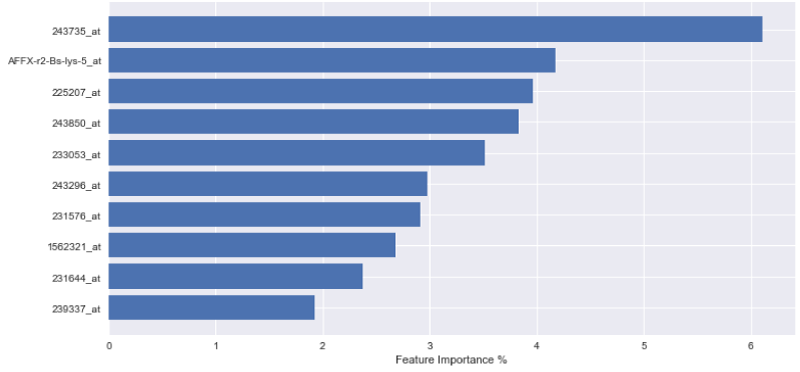
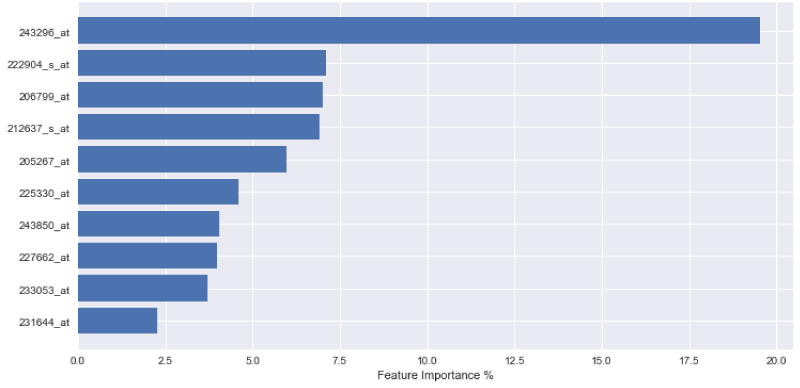
In order to express the weighting of data in the aforementioned decision-making process, the outputs of each variable importance for each tested model are visually displayed in Figures 1-3. The figures demonstrate the most significant 10 array IDs as determined via the use of each respective algorithm. Additionally, the respective contributions of these arrays to the model can be visualized in Table 2, where the array IDs are presented in terms of their corresponding gene name.
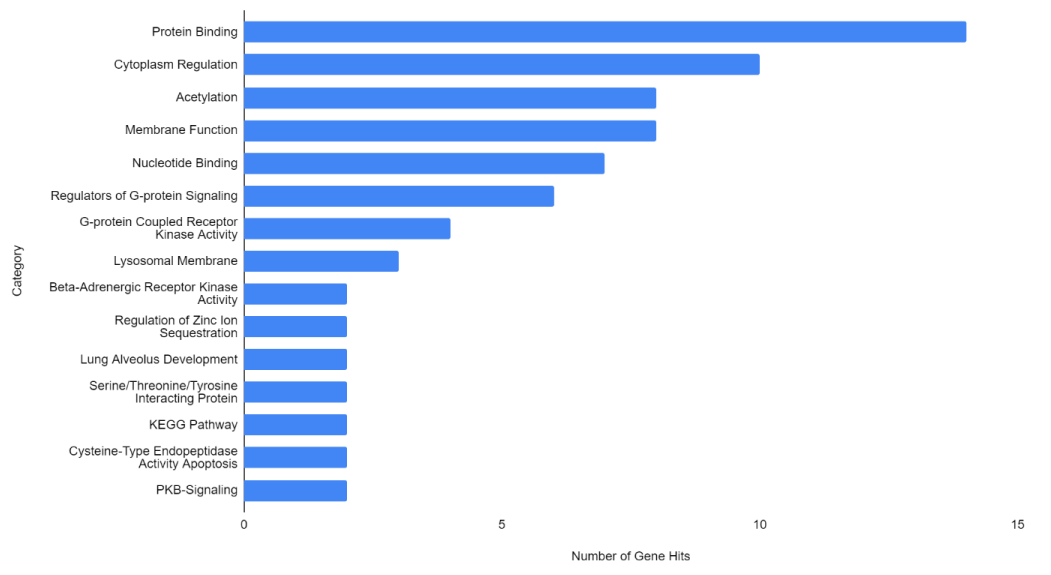
Figure 1-3 represent the variable importance of particular arrays in reaching the decisions within the decision tree models. Figure 4 illustrates the most prevalent biological functions, where differentially expressed genes play a role in the metastatic process.
Table 3 illustrates each algorithm’s predicted top genetic candidates in respective order of priority for metastatic detection. In this table, each variable is listed from highest to lowest importance in metastatic prediction. Common important genes across all of the tested algorithms have been determined to be CD8, PB1, and THP-1, as shown in the table in bold lettering. The prevalence of these expression markers is also indicated. More specifically; differential expression in some of these markers is either present in all cancers, in a variety of different cancers, or is specific to breast cancer or a particular subset of cancers.
This analysis enabled a variety of metastatic biomarkers to be pinpointed, including some unknown genes that have yet to be identified by previous research (indicated by the “N/A” notation). The identifiable genes with the most significant differential expression were discussed below. The most significant genes identified by this analysis are listed and explained in the biological context below.
- One specific target, CD8, also known as CD8A and cluster of differentiation 8, is a transmembrane glycoprotein that was found to have key significance in this analysis. It is a TCR, or T-cell receptor, which facilitates cytotoxic T-cell activity. Upregulation of CD8 has been associated with poor cancer prognosis in recent work by Saleh, et al. [21].
- PB1 (PBRM1 or polybromo 1) is a tumor suppressor gene. Mutations in this gene are ubiquitous across multiple cancer subtypes. This gene encodes an ATP-dependent chromatin-remodeling complex.
- THP-1 or GLI2 is a zinc finger protein referred to as “Glioma-Associated Oncogene Family Zinc Finger 2”. This gene encodes a protein for the zinc finger, which binds DNA and mediates sonic hedgehog signaling (SHH). Disruptions in the SHH pathway have long been associated with cancer and cellular proliferation. The pathway has also been implicated in evolving treatment resistance [22].
- Lastly, the ETNK1 ethanolamine kinase 1 gene encodes the EKI1 kinase protein. This protein is involved in the phosphatidylethanolamine synthesis pathway. Mutations thus affect glycerophospholipid biosynthesis and metabolism.
- Other significant genes were found as well. Two array IDs of interest (1562321_at and 225207_at) were found to correspond to PDK4, or pyruvate dehydrogenase kinase 4. PDK4 is a PDK-BCKDK protein kinase that encodes a mitochondrial histidine kinase protein. When mutated, pyruvate dehydrogenase is no longer regulated, leading to the corresponding dysregulation of glycolysis. PDK4 mutations are ubiquitous in fast-growing cancer cells.
- ELP2, or elongator acetyltransferase complex subunit 2, is another gene of interest. ELP2 encodes a core subunit of the histone acetyltransferase of RNA pol II and is necessary for chromatin remodeling, which is dysregulated in cancer.
- IGF1R encodes the insulin-like growth factor 1 receptor, responsible for binding IGF and exhibiting tyrosine kinase activity. IGF1R is overexpressed in cancers, which confers mutated cells with anti-apoptotic properties.
- TET2, or Tet methylcytosine dioxygenase 2, encodes a gene that catalyzes the conversion of methylcytosine to 5-hydroxymethylcytosine. Gene defects can cause myeloproliferative.
- POU2AF1 encodes a Class 2 Homeobox Associating Factor that associates with OCT1 and OCT2. Defects have been associated with lymphoma.
- SCGB1D2 encodes Secretoglobin Family 1D Member 2, or Prostatein-like Lipophillin B. As a prostatatein analog, the protein encoded by this gene can bind steroid hormones and similar chemotherapeutic agents such as estramustine.
- SYNPO2 produces the protein Synaptopodin 2, which functions in actin bundling and bundling into F-actin. This is necessary for the formation of Z disks and stable autophagocytotic function. ENPP5 is a member of the Ectonucleotide Pyrophosphatase and Phosphodiesterase Family. ENPP5 encodes a type-1 transmembrane glycoprotein that is a prognostic marker in a variety of cancer types.
Lastly, we created a network analysis, as represented in Figure S2, of the output of the gradient boosting results, as this was found to be the most successful model tested within the machine learning analysis. The online GeneMANIA bioinformatics tool was used for this purpose [11]. The GeneMANIA tool searches for information on particular genes and performs network analysis to determine key interactions in the results. When using the GeneMANIA tool, a link showing the interaction between each pair of genes within the target pool is created by analyzing the relationships within the data. The co-expression of transcripts was analyzed, and the interaction links were defined based on previously categorized relationships from data presented in the GeneMANIA Online Tool (https://genemania.org/).
These findings were used to refine a target network for downstream network analysis. Thus, in addition to characterizing an effective tree-based machine learning workflow for metastatic classification of array IDs and determining potential genes at play in early-stage breast cancer metastasis, we have also created a network by looking at the interactions of the differentially expressed genes that were found to play a role in metastasis as represented in Figure S2.
Conclusion
In this study, machine learning decision trees were used to process a clinical genetic expression dataset. In particular, basic decision trees, extremely randomized trees, and gradient boosting trees were compared and assessed in their ability to distinguish between gene expression patterns characteristic of metastatic and nonmetastatic breast cancer.
After model training, it was observed that the gradient boosting tree method was the most powerful algorithm for predicting metastatic potential within the breast cancer dataset. Feature importance analysis enabled array IDs to be narrowed down to a select pool of important arrays that play a significant role in classifying metastasis. Correlated genes and their functions were assessed to understand the broader biological context. It is seen that 243850_at, 233053_at, 231644_at, and 231576_at, are common effective arrays for predicting breast cancer metastasis, indicating that CD8, PB2, THP-1, and ETNK1 are amongst the most significant genes of interest.
In the future, we are planning to extend the study by adding more available next-generation sequencing (NGS) data and using causal inference methods. More research must be conducted to understand what genes correspond to unknown array ID hits that were strongly differentially expressed between metastatic and non-metastatic patients. All code is available on GitHub at http://github.com/melihagraz/ML_Metastatic_Prediction.
Ethical statement
No funding was used for the execution of this research. M.F.B is a consultant for Tersus Life Sciences, LLC. The study was conducted using data from the NCBI GEO database, and appropriate ethical approval and informed consent procedures were followed by the NIH for the collection of this data. No ethics approval was required by the authors for this study.
Consent for publication
Availability of data and materials: In this study, two different publicly available datasets are used. The two datasets are publicly available on NCBI GEO Databank (https://www.ncbi.nlm.nih.gov/geo/). The datasets, called GSE102484 and GSE20685, are merged together to create a single dataset.
- Dillekås H, Rogers MS, Straume O. Are 90% of deaths from cancer caused by metastases? Cancer Med. 2019 Sep;8(12):5574-5576. doi: 10.1002/cam4.2474. Epub 2019 Aug 8. PMID: 31397113; PMCID: PMC6745820.
- Gonzalez H, Hagerling C, Werb Z. Roles of the immune system in cancer: from tumor initiation to metastatic progression. Genes Dev. 2018 Oct 1;32(19-20):1267-1284. doi: 10.1101/gad.314617.118. PMID: 30275043; PMCID: PMC6169832.
- Wang WC, Zhang XF, Peng J, Li XF, Wang AL, Bie YQ, Shi LH, Lin MB, Zhang XF. Survival Mechanisms and Influence Factors of Circulating Tumor Cells. Biomed Res Int. 2018 Nov 1;2018:6304701. doi: 10.1155/2018/6304701. PMID: 30515411; PMCID: PMC6236925.
- Bertucci F, Ng CKY, Patsouris A, Droin N, Piscuoglio S, Carbuccia N, Soria JC, Dien AT, Adnani Y, Kamal M, Garnier S, Meurice G, Jimenez M, Dogan S, Verret B, Chaffanet M, Bachelot T, Campone M, Lefeuvre C, Bonnefoi H, Dalenc F, Jacquet A, De Filippo MR, Babbar N, Birnbaum D, Filleron T, Le Tourneau C, André F. Genomic characterization of metastatic breast cancers. Nature. 2019 May;569(7757):560-564. doi: 10.1038/s41586-019-1056-z. Epub 2019 May 22. Erratum in: Nature. 2019 Aug;572(7767):E7. PMID: 31118521.
- Glare P, Virik K, Jones M, Hudson M, Eychmuller S, Simes J, Christakis N. A systematic review of physicians' survival predictions in terminally ill cancer patients. BMJ. 2003 Jul 26;327(7408):195-8. doi: 10.1136/bmj.327.7408.195. PMID: 12881260; PMCID: PMC166124.
- Secilmis D, Agraz M, Purutcuglu V. Two New Nonparametric Models for Biological Networks, In Hemanchardan K. et al. (editors) Bayesian Reasoning and Gaussian Processes for Machine Learning Applications. CRC Press. 2022.
- Karasu Benyes Y, Welch EC, Singhal A, Ou J, Tripathi A. A Comparative Analysis of Deep Learning Models for Automated Cross-Preparation Diagnosis of Multi-Cell Liquid Pap Smear Images. Diagnostics (Basel). 2022 Jul 29;12(8):1838. doi: 10.3390/diagnostics12081838. PMID: 36010189; PMCID: PMC9406372.
- Deng Y, Lu L, Aponte L, Angelidi AM, Novak V, Karniadakis GE, Mantzoros CS. Deep transfer learning and data augmentation improve glucose levels prediction in type 2 diabetes patients. NPJ Digit Med. 2021 Jul 14;4(1):109. doi: 10.1038/s41746-021-00480-x. PMID: 34262114; PMCID: PMC8280162.
- Acosta JN, Falcone GJ, Rajpurkar P, Topol EJ. Multimodal biomedical AI. Nat Med. 2022 Sep;28(9):1773-1784. doi: 10.1038/s41591-022-01981-2. Epub 2022 Sep 15. PMID: 36109635.
- Cruz JA, Wishart DS. Applications of machine learning in cancer prediction and prognosis. Cancer Inform. 2007 Feb 11;2:59-77. PMID: 19458758; PMCID: PMC2675494.
- Huang MW, Chen CW, Lin WC, Ke SW, Tsai CF. SVM and SVM Ensembles in Breast Cancer Prediction. PLoS One. 2017 Jan 6;12(1):e0161501. doi: 10.1371/journal.pone.0161501. PMID: 28060807; PMCID: PMC5217832.
- Behravan H, Hartikainen JM, Tengström M, Kosma VM, Mannermaa A. Predicting breast cancer risk using interacting genetic and demographic factors and machine learning. Sci Rep. 2020 Jul 6;10(1):11044. doi: 10.1038/s41598-020-66907-9. PMID: 32632202; PMCID: PMC7338351.
- Xiao Y, Wu J, Lin Z, Zhao X. A deep learning-based multi-model ensemble method for cancer prediction. Comput Methods Programs Biomed. 2018 Jan;153:1-9. doi: 10.1016/j.cmpb.2017.09.005. Epub 2017 Sep 14. PMID: 29157442.
- Kadir T, Gleeson F. Lung cancer prediction using machine learning and advanced imaging techniques. Transl Lung Cancer Res. 2018 Jun;7(3):304-312. doi: 10.21037/tlcr.2018.05.15. PMID: 30050768; PMCID: PMC6037965.
- Azzawi H, Hou J, Xiang Y, Alanni R. Lung cancer prediction from microarray data by gene expression programming. IET Syst Biol. 2016 Oct;10(5):168-178. doi: 10.1049/iet-syb.2015.0082. PMID: 27762231; PMCID: PMC8687242.
- Song YY, Lu Y. Decision tree methods: applications for classification and prediction. Shanghai Arch Psychiatry. 2015 Apr 25;27(2):130-5. doi: 10.11919/j.issn.1002-0829.215044. PMID: 26120265; PMCID: PMC4466856.
- Geurts P, Ernst D, Wehenkel L. Extremely randomized trees, Machine Learning. 2006; 63: 3-42.
- Breiman L. Arcing The Edge. The Annals of Statistics. 1998; (3):801-849.
- Friedman JH. Greedy Function Approximation: A Gradient Boosting Machine (PDF). 1999.
- Calza S, Hall P, Auer G, Bjöhle J, Klaar S, Kronenwett U, Liu ET, Miller L, Ploner A, Smeds J, Bergh J, Pawitan Y. Intrinsic molecular signature of breast cancer in a population-based cohort of 412 patients. Breast Cancer Res. 2006;8(4):R34. doi: 10.1186/bcr1517. PMID: 16846532; PMCID: PMC1779468.
- Saleh R, Sasidharan Nair V, Toor SM, Taha RZ, Murshed K, Al-Dhaheri M, Khawar M, Petkar MA, Abu Nada M, Al-Ejeh F, Elkord E. Differential gene expression of tumor-infiltrating CD8+ T cells in advanced versus early-stage colorectal cancer and identification of a gene signature of poor prognosis. J Immunother Cancer. 2020 Sep;8(2):e001294. doi: 10.1136/jitc-2020-001294. PMID: 32948653; PMCID: PMC7511623.
- Carballo GB, Honorato JR, de Lopes GPF, Spohr TCLSE. A highlight on Sonic hedgehog pathway. Cell Commun Signal. 2018 Mar 20;16(1):11. doi: 10.1186/s12964-018-0220-7. PMID: 29558958; PMCID: PMC5861627.
- Kao KJ, Chang KM, Hsu HC, Huang AT. Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: implications for treatment optimization. BMC Cancer. 2011 Apr 18;11:143. doi: 10.1186/1471-2407-11-143. PMID: 21501481; PMCID: PMC3094326.
- Ward R, Sims AH, Lee A, Lo C, Wynne L, Yusuf H, Gregson H, Lisanti MP, Sotgia F, Landberg G, Lamb R. Monocytes and macrophages, implications for breast cancer migration and stem cell-like activity and treatment. Oncotarget. 2015 Jun 10;6(16):14687-99. doi: 10.18632/oncotarget.4189. PMID: 26008983; PMCID: PMC4546497.
- Zhu X, Li S. Retraction Note: TET2 inhibits tumorigenesis of breast cancer cells by regulating caspase-4. Sci Rep. 2019 Mar 28;9(1):5529. doi: 10.1038/s41598-019-39690-5. PMID: 30918283; PMCID: PMC6437154.
- Xia E, Zhou X, Bhandari A, Zhang X, Wang O. Synaptopodin-2 plays an important role in the metastasis of breast cancer via PI3K/Akt/mTOR pathway. Cancer Manag Res. 2018 Jun 18;10:1575-1583. doi: 10.2147/CMAR.S162670. PMID: 30038517; PMCID: PMC6051747.
- Bauer D, Mazzio E, Hilliard A, Oriaku ET, Soliman KFA. Effect of apigenin on whole transcriptome profile of TNFα-activated MDA-MB-468 triple negative breast cancer cells. Oncol Lett. 2020 Mar;19(3):2123-2132. doi: 10.3892/ol.2020.11327. Epub 2020 Jan 22. PMID: 32194710; PMCID: PMC7038999.
- Zafrakas M, Petschke B, Donner A, Fritzsche F, Kristiansen G, Knüchel R, Dahl E. Expression analysis of mammaglobin A (SCGB2A2) and lipophilin B (SCGB1D2) in more than 300 human tumors and matching normal tissues reveals their co-expression in gynecologic malignancies. BMC Cancer. 2006 Apr 9;6:88. doi: 10.1186/1471-2407-6-88. PMID: 16603086; PMCID: PMC1513245.
- Sharma P, Bhattacharyya DK, Kalita J. Disease biomarker identification from gene network modules for metastasized breast cancer. Sci Rep. 2017 Apr 21;7(1):1072. doi: 10.1038/s41598-017-00996-x. PMID: 28432361; PMCID: PMC5430701.
- Thalor A, Kumar Joon H, Singh G, Roy S, Gupta D. Machine learning assisted analysis of breast cancer gene expression profiles reveals novel potential prognostic biomarkers for triple-negative breast cancer. Comput Struct Biotechnol J. 2022 Mar 24;20:1618-1631. doi: 10.1016/j.csbj.2022.03.019. PMID: 35465161; PMCID: PMC9014315.
- Thalor A, Kumar Joon H, Singh G, Roy S, Gupta D. Machine learning assisted analysis of breast cancer gene expression profiles reveals novel potential prognostic biomarkers for triple-negative breast cancer. Comput Struct Biotechnol J. 2022 Mar 24;20:1618-1631. doi: 10.1016/j.csbj.2022.03.019. PMID: 35465161; PMCID: PMC9014315.
- Sun S, Liu J, Zhao M, Han Y, Chen P, Mo Q, Wang B, Chen G, Fang Y, Tian Y, Zhou J, Ma D, Gao Q, Wu P. Loss of the novel mitochondrial protein FAM210B promotes metastasis via PDK4-dependent metabolic reprogramming. Cell Death Dis. 2017 Jun 8;8(6):e2870. doi: 10.1038/cddis.2017.273. Erratum in: Cell Death Dis. 2019 Sep 23;10(10):707. PMID: 28594398; PMCID: PMC5520928.
- Mo D, Li C, Liang J, Shi Q, Su N, Luo S, Zeng T, Li X. Low PBRM1 identifies tumor progression and poor prognosis in breast cancer. Int J Clin Exp Pathol. 2015 Aug 1;8(8):9307-13. PMID: 26464681; PMCID: PMC4583913.
- Zhang H, Zhang N, Liu Y, Su P, Liang Y, Li Y, Wang X, Chen T, Song X, Sang Y, Duan Y, Zhang J, Wang L, Chen B, Zhao W, Guo H, Liu Z, Hu G, Yang Q. Epigenetic Regulation of NAMPT by NAMPT-AS Drives Metastatic Progression in Triple-Negative Breast Cancer. Cancer Res. 2019 Jul 1;79(13):3347-3359. doi: 10.1158/0008-5472.CAN-18-3418. Epub 2019 Apr 2. Erratum in: Cancer Res. 2021 Jun 1;81(11):3145. PMID: 30940661.
- Liu P, Sun Y, Liu S, Niu J, Liu X, Chu Q. SY-707, an ALK/FAK/IGF1R inhibitor, suppresses growth and metastasis of breast cancer cells. Acta Biochim Biophys Sin (Shanghai). 2022 Feb 25;54(2):252-260. doi: 10.3724/abbs.2022008. PMID: 35538024.
- Zhang Q, Li T, Wang Z, Kuang X, Shao N, Lin Y. lncRNA NR2F1-AS1 promotes breast cancer angiogenesis through activating IGF-1/IGF-1R/ERK pathway. J Cell Mol Med. 2020 Jul;24(14):8236-8247. doi: 10.1111/jcmm.15499. Epub 2020 Jun 17. PMID: 32548873; PMCID: PMC7348140.
- Zhou L, Li H, Sun T, Wen X, Niu C, Li M, Li W, Hoffman AR, Hu JF, Cui J. HULC targets the IGF1R-PI3K-AKT axis in trans to promote breast cancer metastasis and cisplatin resistance. Cancer Lett. 2022 Aug 15;548:215861. doi: 10.1016/j.canlet.2022.215861. Epub ahead of print. PMID: 35981570.
- Zeng L, Yu J, Huang T, Jia H, Dong Q, He F, Yuan W, Qin L, Li Y, Xie L. Differential combinatorial regulatory network analysis related to venous metastasis of hepatocellular carcinoma. BMC Genomics. 2012;13 Suppl 8(Suppl 8):S14. doi: 10.1186/1471-2164-13-S8-S14. Epub 2012 Dec 17. PMID: 23282077; PMCID: PMC3535701.
- Cheng M, Bhujwalla ZM, Glunde K. Targeting Phospholipid Metabolism in Cancer. Front Oncol. 2016 Dec 27;6:266. doi: 10.3389/fonc.2016.00266. PMID: 28083512; PMCID: PMC5187387.
- Lau WM, Doucet M, Stadel R, Huang D, Weber KL, Kominsky SL. Enpp1: a potential facilitator of breast cancer bone metastasis. PLoS One. 2013 Jul 5;8(7):e66752. doi: 10.1371/journal.pone.0066752. PMID: 23861746; PMCID: PMC3702501.
- Wu W, Warner M, Wang L, He WW, Zhao R, Guan X, Botero C, Huang B, Ion C, Coombes C, Gustafsson JA. Drivers and suppressors of triple-negative breast cancer. Proc Natl Acad Sci U S A. 2021 Aug 17;118(33):e2104162118. doi: 10.1073/pnas.2104162118. PMID: 34389675; PMCID: PMC8379974.
- Barznegar M, Rahimi K, Mahdavi P, Menbari MN, Darvishi N, Vahabzadeh Z, Hakhamaneshi MS, Andalibi P, Abdi M. Relation between the circular and linear form of the Elongator Acetyltransferase Complex Subunit 3 in the progression of triple-negative breast cancer. Cell Biochem Funct. 2022 Aug;40(6):550-558. doi: 10.1002/cbf.3724. Epub 2022 Jun 20. PMID: 35722999.
- Hogstrand C, Kille P, Ackland ML, Hiscox S, Taylor KM. A mechanism for epithelial-mesenchymal transition and anoikis resistance in breast cancer triggered by zinc channel ZIP6 and STAT3 (signal transducer and activator of transcription 3). Biochem J. 2013 Oct 15;455(2):229-37. doi: 10.1042/BJ20130483. PMID: 23919497; PMCID: PMC3789231.
- Tecalco-Cruz AC, Macías-Silva M, Ramírez-Jarquín JO, Méndez-Ambrosio B. Identification of genes modulated by interferon gamma in breast cancer cells. Biochem Biophys Rep. 2021 Jun 16;27:101053. doi: 10.1016/j.bbrep.2021.101053. PMID: 34189281; PMCID: PMC8220005.
- Wang Y, Dai J, Zeng Y, Guo J, Lan J. E3 Ubiquitin Ligases in Breast Cancer Metastasis: A Systematic Review of Pathogenic Functions and Clinical Implications. Front Oncol. 2021 Oct 22;11:752604. doi: 10.3389/fonc.2021.752604. PMID: 34745984; PMCID: PMC8569917.
- Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD, Morris Q. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010 Jul;38(Web Server issue):W214-20. doi: 10.1093/nar/gkq537. PMID: 20576703; PMCID: PMC2896186.
- Cheng SH, Huang TT, Cheng YH, Tan TBK, Horng CF, Wang YA, Brian NS, Shih LS, Yu BL. Validation of the 18-gene classifier as a prognostic biomarker of distant metastasis in breast cancer. PLoS One. 2017 Sep 8;12(9):e0184372. doi: 10.1371/journal.pone.0184372. PMID: 28886126; PMCID: PMC5590926.

Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
