Global Journal of Clinical Virology
Worldwide remerging of SARS CoV-2 (Severe acute respiratory syndrome coronavirus 2) linked with COVID-19: current status and prospects
Zeshan Haider1*, Maria Javed1 and Numan Yousaf2
2Department of Bioinformatics, Comsats University Islamabad (CUI), Islamabad, Pakistan
Cite this as
Haider Z, Javed M, Yousaf N (2021) Worldwide remerging of SARS CoV-2 (Severe acute respiratory syndrome coronavirus 2) linked with COVID-19: current status and prospects. Glob J Clin Virol 6(1): 012-020. DOI: 10.17352/gjcv.000010A novel coronavirus virus (2019-nCov) emerged in China in December 2019, which posed an International Public Health Emergency in a couple of weeks, and very recently entered World Health Organization (WHO) status as a very high-risk group. The International Committee on Virus Taxonomy (ICTV) called this virus the Extreme Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2), and the disease is known as Coronavirus Disease-19 (COVID-19). The COVID-19 caused nearly 1,913,391 individuals, out of a total of around 88,861,041 confirmed cases affected by this infection until January 8, 2021. This edition offers a brief overview of the most outstanding features and information about the emerging coronavirus infection, the present worldwide scenario and mechanism of illness, replication and dissemination, as well as ongoing progress in the control and management of this disease, which has now spread to more than 100 countries around the world. Note that researchers worldwide and various health agencies are all working together to stop the spread of this virus and avoid any possible pandemic situation that would otherwise endanger millions of people’s lives.
Introduction
The new novel virus is known as the 2019 coronavirus. The International Committee on Virus Taxonomy (ICT V) called it the SARS-CoV-2 virus on February 11, 2020, because it is the highest genome similarity to the SARS CoV [1]. In 1966, they cultivated the coronavirus first described by Tyrell and Bynoe from patients with common colds. The cultivated virus is named a coronavirus (Latin: corona = crown) due to resembles a solar corona, morphology on the outer shell as spikes of surface projection [2]. SARS CoV-2 belong to the Coronaviridae family composed of four types: Alphacoronavirus, Betacoronavirus, Gammacoronavirus and Deltacoronavirus. The novel coronavirus is a positive, unsegmented, and enveloped single-stranded RNA virus. That not only infects humans but also affects a wide range of animals such as camels, bats, avians, rodents, and chiropters [3].
A coronavirus is the cause of neurological and acute respiratory syndrome disorders. Previously, various coronavirus strains, i.e. HCoV-229E, HCoV-HKU1, HCoV-OC43, HCoV-NL63, MERS-CoV and SARS-CoV, have found that infect humans [4]. The length of the entire Genome of SARS CoV-2 is 29881 bp. SARS CoV-2 belongs to the beta coronavirus, infects the human respiratory tract and causes pneumonia similar to MERS and SARS [5]. CoVs continue to undergo recombination and mutation due to their unique replication mechanism, allowing them to continually acclimatize into new hosts and ecological locations [6,7]. The birds and animals served as the reservoir for the emergence of most novel viral strains because of the habit of walking in flocks and their ability to fly long distances. Birds can spread new viruses among themselves and to humans. China’s diverse migratory routes and bird species, including CoVs, can bring several pathogens into the country [6,8]. The lessons learned from these earlier threats to SARS, MERS and existing SARS CoV-2 situations need to be taken into account when formulating strategies to combat these and other emerging and zoonotic pathogens that could pose pandemic threats, thus placing human lives at risk [9]. In this review, we compile the current COVID-19 research insights focusing on enhanced surveillance, current situation and control and precaution of the deadly infections in different countries worldwide.
History and emerging of coronavirus
SARS-Cov, MERS-CoV and the newly discovered SARS-CoV-2 are thought to produce natural selection but not by laboratory development. They are highly pathogenic, causing extreme lower and upper respiratory syndrome extrapulmonary infection. In 1996, the first coronavirus strain HCoV-229E was reported, isolated from the patient’s upper respiratory tract with symptoms including sneezing, sore throat, cough and fever in 10-20 % of cases. [10,11]. Later on, in 1976, another strain of HCoV-OC43 was reported from organ culture, and subsequent serial passage in the brains of suckling mice and symptoms was mostly similar to HCoV-229E [12]. The incubation time of these virus strains was less than two weeks. SARS CoV is the first well-documented human coronavirus, also known as “atypical pneumonia.” SARS CoV belongs to a beta coronavirus family and triggered a pandemic in human history. Many viruses have been identified up to date, but SARS coronavirus shows more similarity, about 98% with the novel SARS-CoV-2 (Figure 1).
Coronavirus spread only after the onset of disease through direct contact of infected people with healthy people. These viruses are epidemic in the human population, causing 15-30 percent of disease in neonates, people with an underlying condition, and the elderly. In particular, SARS coronavirus infected epithelial cells in the lungs. In China’s Guangdong province, the first SARS case tracked back to late 2002, and the epidemic results were about 919 deaths with reported cases of 8,096 and spread across many continents. The incubation period of SARS-Co-V was back, 4 to 7 days, and viral load peak appeared on the 10th day of disease [13,14]. Five additional cases of SARS originating from zoonotic transmission occurred in December 2003-January 2004. However, later on, it is shown that a particular SARS-Co-V-like virus found in bats infected human cells without prior adaptation, which indicates that SARS could reemerge [15]. Infected patients’ symptoms included malaise, chills, headache, fever, and late cough or respiratory distress that increased macrophages, epithelial cell proliferation, and diffuse alveolar damage also occurred in SARS patients [16]. After that, mechanical ventilation and intensive care will require approximately 20-30 percent of patients. Cytokine storm caused damage to other organs, including the gastrointestinal tract, kidney and liver, that can be lethal, especially in patients with immunocompromises. The economy is also negatively affected by SARS, with reduced international travel and domestic demands and an estimated global economic loss of $40 billion [17]. Consequently, the SARS coronavirus outbreak was controlled by prevention in 2003. The virus has not returned since a human coronavirus emerged in 2012 as MERS-CoV, Middle East Respiratory Syndrome.
MERS-CoV was first identified in 2012 from the lungs of a 60-year-old patient who developed acute pneumonia and renal failure in Saudi Arabia. Dipeptidyl peptidase 4 (DPP4) is used as a receptor by MERS-Co-V [18]. Several cases of severe respiratory diseases occurred in 2012 and were reported in Jordan hospital, and three patients also registered in September 2012. According to the European Center for Disease Prevention and Control, this epidemic encourages concern in 2014 that the epidemic has been weakened and was more capable of human to human transmission, a fatality rate of approximately 40 percent with 855 cases and 333 deaths. In 2015 another secondary outbreak occurred in South Africa with 186 confirmed cases; clinical manifestation resembles acute pneumonia SARS. More than 30 percent of patients have gastrointestinal symptoms with diarrhea and vomiting. As of April 26, 2016, 1728 confirmed cases were reported, including 624 deaths and spread across 27 countries. As of February 14, 2020, over 2,500 confirmed laboratory cases with a high case fatality rate of 34.4 percent were registered, making MERS-CoV one of the most damaging viruses known to humans [19,20].
The novel coronavirus SARS Cov-2
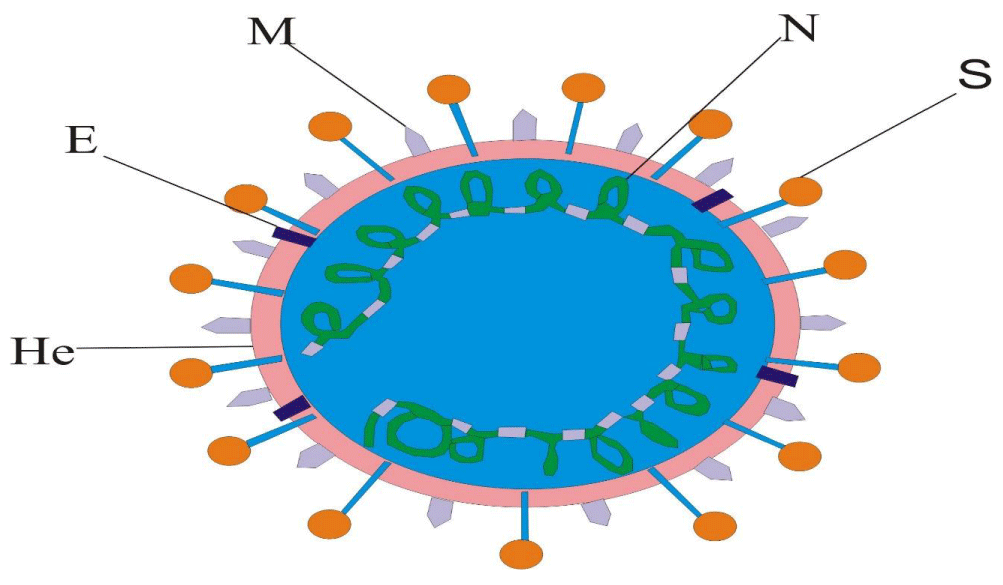
The SARS-CoV-2 is an enveloped, spherical type, single-stranded plus RNA virus. The virus has peplomers made up of glycoprotein projected over the envelope in a crown-like manner (hence called the corona). These spike proteins bind with receptors present in animal and human bodies [21,22]. Changes in receptor binding ligands at the spikes level are responsible for zoonotic spillover and barrier crossing of the species. The high genomic similarities suggest that SARS-CoV-2, which produces COVID-19 in humans, originates from bats as bats function as a natural ancestral host [23,24]. Scanning electron microscopy, transmission electron microscopy and cryoelectron microscopic images of the SARS-CoV-2 structure confirmed the change in spike glycoprotein of SARS coronavirus-2 (Figure 2) [25]. Genomic studies showed that only five nucleotides differed between SARS coronavirus and SARS-CoV-2 and stressed that 2019-nCoV emerged from SARS CoV [26].
Worldwide occurrence of SARS Cov-2
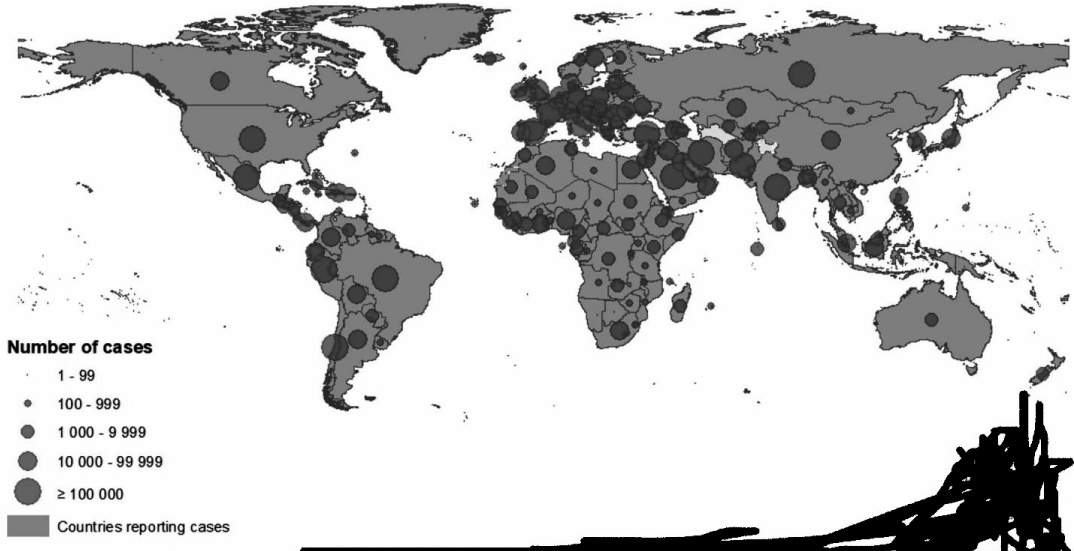
After its initial appearance in Wuhan, China, this recent emerging Co-V was included in the International Emergency Category of Public Health on January 30, 2020 [27,28]. In addition to mainly affecting China, SARS-CoV-2 / COVID-19 has now extended to over 100 countries. Out of cumulative 6960,259 confirmed cases, 401,970 human deaths have documented on June 8, 2020 (Figure 3) [29,30]. The WHO has established a very high-risk community of COVID-19 due to its rapid spread in several countries over a short period (Figure 4). Few of the reports have mentioned the potential for likely pandemic risks and threats to the bloom. We are appearing in increasingly the cases of COVID-19, alarming us to make substantial efforts to monitor the spread of this widespread virus among the world’s population by adopting effective prevention and control measures and formulating global approaches and updated strategies with prospects [9,30,31].
Coronavirus’s cycle for entering in host cell
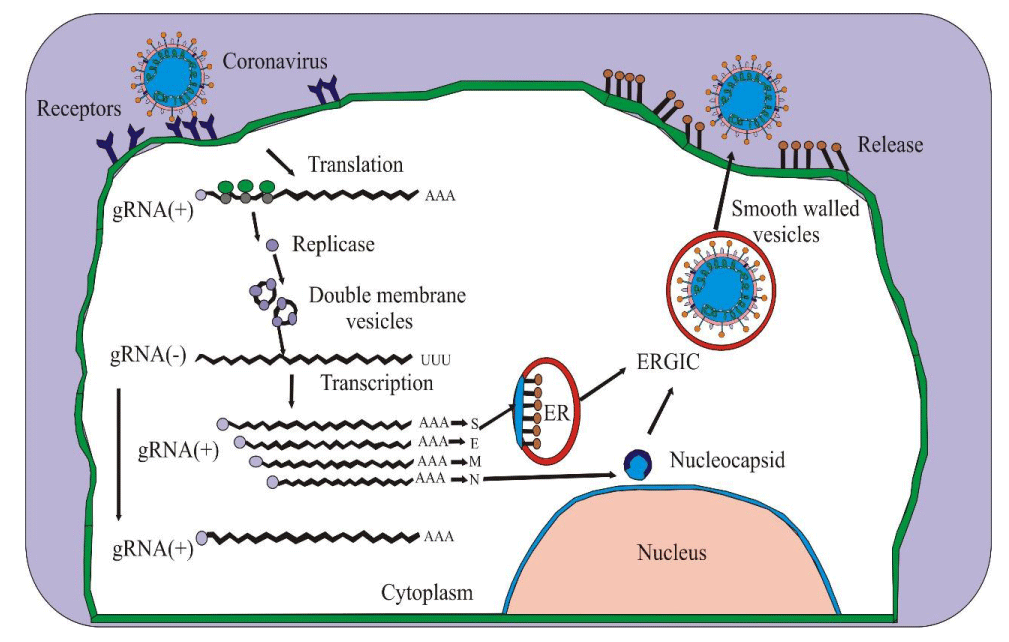
All coronavirus has a unique gene area in open reading frame 1(ORF1). These genes encode replication proteins, spike formation, nucleocapsid. The first important step in coronavirus infection is to bind the glycoprotein S spike protein to the host human angiotensin-converting enzyme two receptors (ACE2) (Bertram, et al. 2011). After the virus entered the host cell cytosol followed by the fusion of viral and cellular membranes, S protein’s cleavage by cathepsin, TMPRRS2 or another protease into S1 and S2 has occurred. The first cleavage is necessary to separate the RBD and the fusion domain. The second is to reveal the fusion peptide and ultimately release the viral genome into the cytoplasm (Figure 3) [21]. The next step is to translate the genes from the virion genomic RNA into replicase.
The replicase genes encode two large Open Read Frames (ORFs) rep1a and rep1b, expressing two pp1a and pp1ab polyproteins using a slippery sequence (5′-UUUAAAC-3′) and a pseudoknot RNA. The pseudoknot RNA induces the rep1a to rep1b ORF by ribosomal frameshifting (Tripp and Tompkins 2018). These structural proteins make up the network of replicate-transcription RNA that complex located in the intracellular membrane to the rough endoplasmic and the -sense RNA strand synthesized by replication and transcription. The -sense RNA strand is used as a template for producing the full length + sense RNA strand during reproduction. Subgenomic RNA codes for all structural proteins formed by discontinuous transcription during the negative RNA strand synthesized by combining the varying length of the genome’s three primary ends with the five primary leader sequence necessary for translation. Afterwards, the subgenomic-sense RNA transcribed into a + sense RNA strand. After forming viral and structural protein components, the structural proteins and the viral genome assemble into the nucleocapsid and viral envelope in the intermediate endoplasmic compartment of Golgi and release transformed virion from the infected cell (Figure 5) (Hussain, et al. 2005). These are transmitted to healthy people by the infected person’s cough, sneezing or indirect contact. It mainly affects the lungs’ alveolar epithelial cells because the receptor for this virus is present primarily in that region.
Sign and symptoms of COVID-19 patients
The symptoms of COVID-19 infection follow an incubation period of ~5.2 days [27]. The duration of signs of infection with diseases to death ranged from 6 to 41 days with a mean of 14 days, and this duration depends on the patient’s immune system and age. The infection rate is> 70 years of age relative to 70 years of age [6]. Initial symptoms of COVID-19 include dry cough, muscle ache, headache, chest pain, diarrhea, vomiting, nausea, rhinorrhea, sore throat, dizziness, and shortness of breath [32]. Many patients developed hypoxemia and dyspnea one week later from the disease infection. In contrast, patients in extreme cases continued developing acute respiratory syndrome, metabolic acidosis, septic shock and coagulopathy. Patients suffering from acute fever or respiratory symptoms are screened for early diagnosis of virus attack. The study report taken at the end of December 2019 shows the percentage of coronavirus symptoms with 76% dry cough, 3% diarrhea, 55% dyspnea, 98% fever, and 8% of patients with ventilation support [33,34]. A 2012 study showed that patients suffering from MERS-CoV diseases also had the same symptoms as COVID-19 included fever 98%, dyspnea 55%, dry cough 47% and 80% required for ventilation support [35]. Guan, et al. reported 1099 cases of infection with CoV–2019. They found the most common symptoms to be 67.7 percent cough and 87.9 percent fever, while 5.0 percent vomiting and 3.7 percent diarrhea are rare. The chest CT image of the SARS-Cov-2 infected patients shows 96 percent abnormalities and 82.1 percent lymphopenia [32].
Diagnosis the victims of COVID-19
Rapid diagnosis of SARS-CoV-2 is widely preferred for molecular tools [36]. Serological testing at the height of the outbreak is of little use, though serum samples of recovered patients could be taken to know the IgG titer. In severely infected patients, Computed Tomography (CT) and X-Ray techniques can help observe pulmonary pneumonia lesions in the lungs in correlation with clinical symptoms to show the COVID-19 picture [37]. The detection of Viral Nucleic Acid (VNA) is essential for diagnosing exposed but asymptomatic carriers. It is possible to detect the viral RNA using a pharyngeal swab to avoid transmission and spread risk [5]. Most popularly, RT-PCR (RRT-qPCR) is performed in real-time over respiratory secretions to identify viral RNA within a short time [38]. Researchers have developed a diagnostic technique for rapidly detecting COVID-19 by Reverse Transcriptional Loop-Mediated Isothermal Amplification (RT-LAMP). This isothermal COVID-19 detection method based on LAMP, referred to as iLACO. In this technique, six primers are used to amplify a fragment of the ORF1ab gene, and phenol red is used as a pH indicator when the amplification changes colour from pink to light yellow. At the same time, it remains rosy in negative cases [39]. Also, multiple reference laboratories are advancing sequencing of the complete genome from the RRT-PCR’s positive isolates.
Treatments against SARS-Cov-2 infection
Besides, the lipid solvent can effectively inactivate these viruses. Except for chlorhexidine, the solvent includes ethanol, ether (75%), peroxyacetic acid, chlorine-containing disinfectant and chloroform. Based on previous experience in combating the SARS-CoV and MARS-CoV epidemic, we learn some treatment strategies against coronavirus. Antiviral therapy is widely used in clinical practice [40]. Remdesivir (GS-5734) is an analogue prodrug of 1′cyano-substituted adenosine nucleotide, and it displays intense antiviral activity against the RNA virus. The first COVID-19 cases in the US were treated successfully with remdesivir drug [41]. Chloroquine has been used for treating malaria for many years. Many mechanisms have been investigated: in several viruses, chloroquine can inhibit the pH-dependent replication steps with a potent effect on SARS-CoV’s spread and infection [42].
It also functions as an ac class of autophagy inhibitor, suppressing the release/production of TNF-α and IL-6 and interfering with the glycosylation of SARS-CoV cell receptors and serving in Vero E6 cells at both the entry and post-entry stages of the COVID-19 infection. The use of a combination of chloroquine and Remdesivir can effectively inhibit In vitro SARS-CoV-2 [43]. The combination of ritonavir and lopinavir antiretroviral drugs improved patients’ clinical condition with SARS-CoV, and it may be an option to treat COVID-19 infections.
Doctors recently isolated COVID-19 blood plasma from recovered patients in Shanghai and injected it into an infected person, who later showed positive results with rapid recovery within 24 hours, accompanied by reduced viral loads, inflammation and improved blood oxygen saturation [44]. They extracted a monoclonal antibody (CR3022), which binds on RBD spikes with SARS-CoV. It has potential for COVID-19 treatment by combination or itself [44,45]. But in addition to its advantages, it has disadvantages because antibodies can fight against any invading pathogen to over-stimulate the cytokine release syndrome that is potentially life-threatening toxicity [46].
For thousands of years, Traditional Chinese Medicines (TCM) have been used to treat several diseases, e.g. Shu Feng Jie Du capsules and Lian Hua Qing Wen capsules. These operated as an effective alternative treatment since no such unique and successful therapy has developed for COVID-19. Cure rate recorded in China using TCM for therapy in many cities such as Gansu (63.7%), Ningxia [47].
Nonetheless, several drugs are under study, including other antiretrovirals, such as remdesivir, antivirals such as oseltamivir and other therapies, including chloroquine and even indomethacin. Researchers are making great efforts to design and produce effective COVID-19 vaccines, which could take some time [48,49]. In this context, effective management of COVID-19 pneumonia by active prevention and scientific control is of utmost importance following the national and international guidelines developed [50].
Prevention and control of COVID-19
It is crucial to avoid the possibility of spark (originating at the new site) and spread (transmission between susceptible and infected). Both to prevent the transformation of the COVID-19 outbreak into a pandemic and, for this reason, intensive monitoring should determine the trend of emerging zoonotic epidemics [51,52]. The protection of individuals and the community must both be robust. The World Health Organization (WHO), the Center for Disease Control and Prevention ( CDC) and the Food and Agriculture Organization (FAO) have issued instruction and COVID-19 containment strategies to be followed by ordinary people, clinicians, travellers and infected patients to prevent transmission to a healthy population [29,53-55]. It is advisable to share awareness programs through social networking sites and platforms and follow intensive epidemiological surveillance to notify WHO of any new cases (symptomatic and asymptomatic) of COVID-19 [4,56]. Bat CoVs should screen epidemiologically globally to have a data sheet that will be a pathfinder for newly emerging and reemerging zoonotic pathogens [57].
Extensive interventions, focus, reduction of transmission and efforts to protect populations, including health care providers, older adults and children, prevent existing outbreaks of COVID-19 infection by governments. Healthcare staff and physicians were healthcare workers at high risk of COVID-19 transmission in 2002 of the SARS outbreak 21 percent of those affected. In China, nearly 3,387 health care staff have been contaminated, and six have died [58]. Most of the COVID-19 death cases occur in an orderly manner due to their weak immune system, which allows for faster progression of viral infection. Guidelines published for people against COVID-19 disease are to reduce social activity, avoid crowded areas, postpone non-essential travel, the distance each other, cough in sleeve/tissue rather than hand, wash hands with sanitizer or alcoholic soap at least 20 seconds after every 15-20 minutes on a routine basis, wear gloves and surgical masks or N95 [59]. The N95 respirator mask can protect against the virus’s inhalation as small as 10 to 80 nm. The virus particle stays on the material for a few hours. Therefore, do not touch hazardous materials, do decontamination of rooms, surfaces, and equipment regularly. The US and other countries, including China, have implemented paramount prevention and screening to control the virus’s future spread [60]. Besides all the one health approach, due attention is also needed to prevent and control this disease and other likely future epidemics [61].
Complex emergencies of COVID-19: Impact, management, challenges and experience in zhuhai, china
It can be concluded from China’s experience that establishing strict rules for controlling or preventing the COVID-19 pandemic by powerful collaboration and organization, strict isolation and disinfection measures, quarantine, availability of temporary hospitals, adequate healthcare workers and highly infected areas becomes locked down. After all, by the government and medical workers’ continuous efforts, China took control of the COVID-19 infection. On March 18, 2020, all the infected patients of Wuhan’s had been discharged after recovery. China, South Korea, and Japan also adopted these strict measures to control the COVID-19 epidemic and become successful in achieving their goal.
Zhuhai is a tourist destination. About 1.89 million citizens of Zhuhai are migrants who work and live there but return for the spring festival back to their hometown, many from Hubei Province on February 2, 2020. The COVID-19 first three cases were diagnosed in Zhuhai on January 20, 2020, who had travelled from Wuhan. After the spring festival, the number is increasing steadily, by 76 patients were diagnosed on February 7, 2020. The Government of Zhuhai took action, and all religious activities were cancelled to avoid gathering, wear a mask, and reduce public transport to <5% on February 7, 2020. From Hubei Province, the Zhuhai citizen who is returning was isolated for 14 days in hospitals. And patient whose body temperature was>37.3°C sent to the clinic for further examination. Two hospital wards are specifically reserved for emergency isolation. The cleaning staff’s protective clothes have improved, pens, mobile phones, stethoscopes and spectacles were disinfected [59].
A lesson to be learned from china and by twain to control pandemic by all other countries
The China experience can be summed up in these points. (1) strict isolation has taken. During the early phase of the epidemic, the government should take strong actions by locking down the infected areas, and the larger areas can be quarantine. (2) Temporary hospital establishment. The establishment of a temporary hospital is vital for patients who have mild COVID-19 symptoms where they can have isolated, so the other family members cannot be infected. It is important to keep healthy people away from infected patients. When no vaccine is available, isolation is the effective way to eliminate COVID-19 from further spreading. COVID-19. (3) Experts with knowledge of COVID-19. Knowledge about COVID-19 in experts is vitally important. In fact, from China to other countries, the experts are drafted. These experts have experience with COVID-19. (4) significant communication. In controlling the pandemic, transparency and communication play an important role in controlling pandemics [62].
The lesson for controlling and preventing the pandemic could also be learned from Taiwan province. As soon as China reported the first case of COVID-19 on December 13, 2019, Twain quickly applies policy decisions and began health checking on broad flights coming from Wuhan. Rapid measurements helped out to isolates COVID-19 first case on January 20, 2020. before February, Twain Province developed 4-hour kits and isolated COVID-19 two strains. During the COVID-19 outbreak, the Taiwan Province applied strategies within the 50 days that included human services, healthcare systems, inter-jurisdictional and intergovernmental funding and adequate funding to ensure response capacity and emergency preparedness [63]. In those countries where strict measures are not taken, they are still affected by the epidemic because of not wearing masks, social distancing, and unhygienic conditions.
Conclusions and future prospects
Wide-ranging scientists, researchers and numerous health agencies workdays and nights with a great deal of effort to stop further transmission and spread of SARS-CoV-2. They are implementing strict surveillance, intervention approaches, improved prevention and control policies, and combating COVID-19 by developing effective vaccines and therapies to prevent any pandemic situation that may arise. Although many aspects derived from the research still need to be developed, the case with many elements in certain age groups facing the COVID-19 pandemic is not clear in pediatrics year after year [64,65]. Lastly, one health approach would play an essential role in the future fight against COVID-19 and against such diseases. Prevention is not a silver bullet, but yes, the world needs global solutions to stop a pandemic or minimize it [66-72].
All the authors recognize their respective Institutes and Universities and thank them.
Funding
The anthology is a review article written, analyzed and conceived by its authors and did not require any significant funding.
Author’s contributions
All authors contributed substantially to the design, design, analysis, and interpretation of the data, checking and approving the final version of the manuscript, and agreeing to be accountable for its content.
Disclosure statement
The Authors announce that there are no commercial or financial relationships which could lead to a possible conflict of interest in any way.
- Gorbalenya AE, Baker SC, Baric R, Groot RJD, Drosten C, et al. (2020) Severe acute respiratory syndrome-related coronavirus: The species and its viruses–a statement of the Coronavirus Study Group. Link: http://bit.ly/2OzAN2q
- Tyrrell DAJ, Bynoe ML (1966) Cultivation of viruses from a high proportion of patients with colds. Lancet 1: 76-77. Link: http://bit.ly/2N9ixg4
- Lim XF, Lee CB, Pascoe SM, How CB, Chan S, et al. (2019) Detection and characterization of a novel bat-borne coronavirus in Singapore using multiple molecular approaches. J Gen Virol 100: 1363-374. Link: http://bit.ly/3rzNclF
- Su S, Wong G, Shi W, Liu J, Lai AC, et al. (2016) Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol 24: 490-502. Link: http://bit.ly/2OarTZy
- Hu Z, Song C, Xu C, Jin G, Chen Y, et al. (2020) Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Sci China Life Sci 63: 706-711. Link: https://bit.ly/38lxJhn
- Wang W, Tang J, Wei F (2020) Updated understanding of the outbreak of 2019 novel coronavirus (2019‐nCoV) in Wuhan, China. J Med Virol 92: 441-447. Link: http://bit.ly/2OlC3pY
- Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, et al. (2012) Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol 86: 3995-4008. Link: http://bit.ly/3qz8kqE
- Mackenzie JS, Jeggo M (2013) Reservoirs and vectors of emerging viruses. Curr Opin Virol 3: 170-179. Link: http://bit.ly/2OgKjHX
- Rodriguez-Morales AJ, Bonilla-Aldana DK, Balbin-Ramon GJ, Rabaan AA, Sah R, et al. (2020) History is repeating itself: Probable zoonotic spillover as the cause of the 2019 novel Coronavirus Epidemic. Infez Med 28: 3-5. Link: http://bit.ly/3cgq5pH
- Hamre D, Procknow JJ (1966) A new virus isolated from the human respiratory tract. Proc Soc Exp Biol Med 121: 190-193. Link: http://bit.ly/2OkOum7
- McIntosh K, Dees JH, Becker WB, Kapikian AZ, Chanock RM (1967) Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc Natl Acad Sci U S A 57: 933-940. Link: http://bit.ly/3qzgqzw
- Tyrrell DAJ, Cohen S, Schilarb JE (1993) Signs and symptoms in common colds. Epidemiol Infect 111: 143-156. Link: http://bit.ly/3qwiacX
- Cheng VC, Lau SK, Woo PC, Yuen KY (2007) Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clinical microbiology reviews 20: 660-694. Link: https://bit.ly/2OIHcZm
- Peiris JSM, Lai ST, Poon LLM, Guan Y, Yam LYC, et al. (2003) Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361: 1319-1325. Link: http://bit.ly/30tLoi6
- Menachery VD, Yount BL, Debbink K, Agnihothram S, Gralinski LE, et al. (2015) A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med 21: 1508-1513. Link: http://bit.ly/38qMjEn
- To KK, Hung IF, Chan JF, Yuen KY (2013) From SARS coronavirus to novel animal and human coronaviruses. J Thorac Dis 5: S103-S108. Link: http://bit.ly/3eqPf7B
- Oberholtzer K, Sivitz L, Mack A, Lemon S, Mahmoud A, et al. (2004) Learning from SARS: preparing for the next disease outbreak: workshop summary. National Academies Press. Link: http://bit.ly/30wAHLA
- Lu L, Liu Q, Du L, Jiang S (2013) Middle East respiratory syndrome coronavirus (MERS-CoV): challenges in identifying its source and controlling its spread. Microbes Infect 15: 625-629. Link: http://bit.ly/38rOoQl
- Hilgenfeld R, Peiris M (2013) From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antiviral Res 100: 286-295. Link: http://bit.ly/3t463pd
- Peck KM, Burch CL, Heise MT, Baric RS (2015) Coronavirus host range expansion and Middle East respiratory syndrome coronavirus emergence: biochemical mechanisms and evolutionary perspectives. Annu Rev Virol 2: 95-117. Link: http://bit.ly/3cf2uWt
- Xia S, Zhu Y, Liu M, Lan Q, Xu W, et al. (2020) Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol Immunol 17: 765-767. Link: https://go.nature.com/3rB3ivb
- Xiao K, Zhai J, Feng Y, Zhou N, Zhang X, et al. (2020) Isolation and characterization of 2019-nCoV-like coronavirus from Malayan pangolins. BioRxiv. Link: http://bit.ly/3eq37iD
- Malik YS, Sircar S, Bhat S, Sharun K, Dhama K, et al. (2020) Emerging novel coronavirus (2019-nCoV)—current scenario, evolutionary perspective based on genome analysis and recent developments. Vet Q 40: 68-76. Link: http://bit.ly/3l0smJw
- Riccucci M (2012) Bats as materia medica: an ethnomedical review and implications for conservation. Vespertillio 16: 249-270. Link: https://bit.ly/3ceyfPy
- Song W, Gui M, Wang X, Xiang Y (2018) Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog 14: e1007236. Link: http://bit.ly/2OIHKyo
- Benvenuto D, Giovanetti M, Ciccozzi A, Spoto S, Angeletti S, et al. (2020) The 2019‐new coronavirus epidemic: evidence for virus evolution. Journal of medical virology 92: 455-459. Link: https://bit.ly/38sUBvz
- Li Q, Guan X, Wu P, Wang X, Zhou L, et al. (2020) Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. New England Journal Medicine. Link: https://bit.ly/3rAiCrU
- Liu SL, Saif L (2020) Emerging viruses without borders: the Wuhan coronavirus. Viruses 12: 130 Link: http://bit.ly/3rB3zhH
- Coronavirus Novel 2019a. "Situation Report-10-30 January 2020." Link: https://bit.ly/3t46ujl
- Azamfirei R (2020) The 2019 novel coronavirus: a crown jewel of pandemics?. J Crit Care Med (Targu Mures) 6: 3-4. Link: http://bit.ly/3eCtMJl
- Rodríguez-Morales AJ, MacGregor K, Kanagarajah S, Patel D, Schlagenhauf P (2020) Going global–Travel and the 2019 novel coronavirus. Travel Med Infect Dis 33: 101578. Link: http://bit.ly/3cjV04j
- Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, et al. (2020) Clinical characteristics of coronavirus disease 2019 in China. New England journal of medicine 382: 1708-1720. Link: https://bit.ly/3t46VtZ
- Calisher C, Carroll D, Colwell R, Corley RB, Daszak P, et al. (2020) Statement in support of the scientists, public health professionals, and medical professionals of China combatting COVID-19. Lancet 395: e42-e43. Link: https://bit.ly/3kZQFr4
- Huang C, Wang Y, Li X, Ren L, Zhao J, et al. (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395: 497-506. Link: http://bit.ly/2OEQvsW
- Yi Y, Lagniton PN, Ye S, Li E, Xu RH (2020) COVID-19: what has been learned and to be learned about the novel coronavirus disease. Int J Biol Sci 16: 1753-1766. Link: http://bit.ly/3qx0VZ7
- Zhang N, Wang L, Deng X, Liang R, Su M, et al. (2020) Recent advances in the detection of respiratory virus infection in humans. J Med Virol 92: 408-417. Link: http://bit.ly/3sZcWYO
- Xu Z, Shi L, Wang Y, Zhang J, Huang L, et al. (2020) “Lancet Respir. Med.” Link: http://bit.ly/3qGBHZr
- Meyer B, Müller MA, Corman VM, Reusken CB, Ritz D, et al. (2014) Antibodies against MERS coronavirus in dromedary camels, United Arab Emirates, 2003 and 2013. Emerg Infect Dis 20: 552-559. Link: http://bit.ly/2PGvkHw
- Yu L, Wu S, Hao X, Dong X, Mao L, et al. (2020) Rapid detection of COVID-19 coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. Clin Chem 66: 975-977. Link: http://bit.ly/3buJDaO
- Zumla A, Chan JF, Azhar EI, Hui DS, Yuen KY (2016) Coronaviruses—drug discovery and therapeutic options. Nat Rev Drug Discov 15: 327-347. Link: http://bit.ly/38pBH8N
- Anderson AS, Scully IL, Buurman ET, Eiden J, Jansen KU (2016) Staphylococcus aureus clumping factor a remains a viable vaccine target for prevention of S. aureus infection. MBio 7: e00225. Link: http://bit.ly/3v7uBPY
- Golden EB, Cho HY, Hofman FM, Louie SG, Schönthal AH, et al. (2015) Quinoline-based antimalarial drugs: a novel class of autophagy inhibitors. Neurosurg Focus 38: E12. Link: http://bit.ly/3bBU5gR
- Derebail VK, Falk RJ (2020) ANCA-associated vasculitis—refining therapy with plasma exchange and glucocorticoids. N Engl J Med 382: 671-673. Link: http://bit.ly/3t6nyVM
- Derebail VK, Falk RJ (2020) ANCA-associated vasculitis—refining therapy with plasma exchange and glucocorticoids. Link: http://bit.ly/3t6nyVM
- Tian X, Li C, Huang A, Xia S, Lu S, et al. (2020) Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect 9: 382-385. Link: http://bit.ly/38oyVk2
- Lee DW, Gardner R, Porter DL, Louis CU, Ahmed N, et al. (2014) Current concepts in the diagnosis and management of cytokine release syndrome. Blood 124: 188-195. Link: http://bit.ly/3eq4a1W
- Lu H (2020) Drug treatment options for the 2019-new coronavirus (2019-nCoV). Biosci Trends 14: 69-71. Link: http://bit.ly/2ODYaI6
- Singh SP, Kumar R, Kumari P, Mitra A (2013) An alternate protocol for establishment of primary caprine fetal myoblast cell culture: an in vitro model for muscle growth study. In Vitro Cell Dev Biol Anim 49: 589-597. Link: http://bit.ly/2OhkN5m
- Dhama K, Chakraborty S, Kapoor S, Tiwari R, Kumar A, et al. (2013) One world, one health-veterinary perspectives. Adv Anim Vet Sci 1: 5-13. Link: https://bit.ly/3bxkSuB
- Gao ZC (2020) Efficient management of novel coronavirus pneumonia by efficient prevention and control in scientific manner. Zhonghua jie he he hu xi za zhi 43: E001. Link: http://bit.ly/3vcJY9K
- Ellwanger JH, Chies JAB (2018) Zoonotic spillover and emerging viral diseases–time to intensify zoonoses surveillance in Brazil. Braz J Infect Dis 22: 76-78. Link: http://bit.ly/2OHfwDP
- Ruiz-Saenz J, Bonilla-Aldana K, Suárez JA, Franco-Paredes C, Vilcarromero S, et al. (2019) Brazil burning! What is the potential impact of the Amazon wildfires on vector-borne and zoonotic emerging diseases?-A statement from an international experts meeting. Travel Med Infect Dis 31: 101474. Link: http://bit.ly/3l0QSdD
- Ierardi AM, Wood BJ, Gaudino C, Angileri SA, Jones EC, et al. (2020) How to handle a COVID-19 patient in the angiographic suite. Cardiovasc Intervent Radiol 43: 820-826. Link: http://bit.ly/3qruknu
- Jernigan DB, CDC COVID-19 Response Team (2020) Update: Public Health Response to the Coronavirus Disease 2019 Outbreak—United States. MMWR. Link: http://bit.ly/3l5EBob
- World Health Organization (2017) Communicating risk in public health emergencies: a WHO guideline for emergency risk communication (ERC) policy and practice. World Health Organization. Link: https://bit.ly/38mGNm8
- Anneliese D, Sam M, Emilie K, Raman P, Annelies WS, et al. (2020) The pandemic of social media panic travels faster than the COVID-19 outbreak. J Travel Med taaa031. Link: http://bit.ly/2OBApQY
- Wong AC, Li X, Lau SK, Woo PC (2019) Global epidemiology of bat coronaviruses. Viruses 11: 174. Link: http://bit.ly/3rAa9F5
- Chang D, Xu H, Rebaza A, Sharma L, Cruz CSD (2020) Protecting healthcare workers from subclinical coronavirus infection. Lancet Respir Med 8: e13. Link: http://bit.ly/2OzELrQ
- Jin YH, Cai L, Cheng ZS, Cheng H, Deng T, et al. (2020) A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version). Military Medical Research 7: 1-23. Link: https://bit.ly/3v51AnR
- Carlos WG, Dela Cruz CS, Cao B, Pasnick S, Jamil S (2020) Novel Wuhan (2019-nCoV) Coronavirus. Am J Respir Crit Care Med 201: P7-P8. Link: http://bit.ly/2OC2ffV
- Daszak P, Olival KJ, Li H (2020) A strategy to prevent future epidemics similar to the 2019-nCoV outbreak. Biosaf Health 2: 6-8. Link: http://bit.ly/3vb4N5G
- Mackolil J, Mackolil J (2020) Addressing psychosocial problems associated with the COVID-19 lockdown. Asian J Psychiatr 51: 102156. Link: http://bit.ly/3cwWXLd
- Lin C, Braund WE, Auerbach J, Chou JH, Teng JH, et al. (2020) Policy decisions and use of information technology to fight coronavirus disease, Taiwan. Emerg Infect Dis 26: 1506-1512. Link: http://bit.ly/2OIKuMc
- Arteaga-Livias FK, Rodriguez-Morales AJ (2020) La comunicación científica y el acceso abierto en la contención de enfermedades: El caso del coronavirus novel 2019 (2019-nCoV). Revista Peruana de Investigación en Salud 4: 07-08. Link: http://bit.ly/2Oa3hA9
- Fang F, Luo XP (2020) Facing the pandemic of 2019 novel coronavirus infections: the pediatric perspectives. Zhonghua er ke za zhi 58: E001. Link: http://bit.ly/3t1MbTJ
- Reeves B, Robinson T, Ram N (2020) Time for the human screenome project. Nature 577: 314-317. Link: http://bit.ly/3cmpSS5
- Chen L, Liu W, Zhang Q, Xu K, Ye G, et al. (2020) RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg Microbes Infect 9: 313-319. Link: http://bit.ly/2N1gWIW
- (2019) “Situation Report, 22.” World Health Organization. Link: http://bit.ly/3ryxDdH
- Jin H, Liu J, Cui M, Lu L (2020) Novel coronavirus pneumonia emergency in Zhuhai: impact and challenges. J Hosp Infect 104: 452-453. Link: http://bit.ly/3qsatVa
- Rodriguez-Morales AJ, Gallego V, Escalera-Antezana JP, Méndez CA, Zambrano LI, et al. (2020) COVID-19 in Latin America: The implications of the first confirmed case in Brazil. Travel Med Infect Dis 35: 101613. Link: http://bit.ly/3qA3dql
- Wang X, Rong W (2020) Predicting acute respiratory infection in Chinese healthy individuals: A effective way of patient care. Journal of King Saud University-Science 32: 1065-1068. Link: http://bit.ly/3l1o8kO
- Yang Z, Mao G, Liu Y, Chen YC, Liu C, et al. (2013) Detection of the pandemic H1N1/2009 influenza A virus by a highly sensitive quantitative real-time reverse-transcription polymerase chain reaction assay. Virol Sin 28: 24-35. Link: http://bit.ly/3l0w2ei
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
