Global Journal of Biotechnology and Biomaterial Science
Key Factors for Successful Protein Purification and Crystallization
Eijaz Ahmed Bhat1*≠, Mohnad Abdalla2 ≠and Irfan A Rather3*
Irfan A. Rather, Applied Microbiology and Biotechnology, Yeungnam University, South Korea, E-mail: rather@ynu.ac.kr
Cite this as
Bhat EA, Abdalla M, Rather IA (2018) Key Factors for Successful Protein Purification and Crystallization. Glob J Biotechnol Biomater Sci 4(1): 001-007. DOI: 10.17352/gjbbs.000010Protein purification and crystallization problems have been noticed for many years. The strategies developed for protein solubility and crystallization have improved the protein production and provide high-resolution crystals for structural studies. The protein solubility is achieved by the Site-directed mutagenesis that generates the hydrophobic to hydrophilic mutations. However, the purity and rate of crystallization success still needs to be improved. In this review, the key factors such as the expression system, Affinity –Tags, solubility, reducing or oxidizing environment, denaturing agents, concentration of precipitant, concentration of protein, ionic strength, isoelectric point and pH, temperature, additives, ligands, presence of substrates, coenzymes, mutation, that affect both protein purification and crystallization are discussed in detail. The aim of this review is to have a profound discussion on these key factors and analyze them in relation to both aspects; purification and crystallization and provide a fruitful advice for boosting the production rate of protein and crystallization effectively.
Introduction
Proteins are large bio-molecules present in their native state which perform the various biological functions. Proteins are often associated with other proteins which lead them to form an oligomeric complex. The polydispersity of the protein sample or binding of ligand or size of proteins can be characterized by using the Dynamic Light Scattering (DLS) [1]. After isolation, proteins are insoluble in their innate state and must be denatured to solubilization. In many cases, recombinant proteins produced in Escherichia coli (E. coli) are partially folded intermediates which form the aggregates (insoluble form) known as inclusion bodies. The extracted proteins from the inclusion bodies are used to study the conformation changes by using denaturants. This is also achieved by using the mild conditions to recover the functional active form (soluble) by using different concentrations of Chaotropic agents like urea and guanidine hydrochloride (GdnHcL) which significantly improve the yield of a recombinant protein [2,3]. The chaotropic agents increase the entropy by interfering with the noncovalent forces like hydrogen bonding, Vander wall forces, and hydrophobic effects.
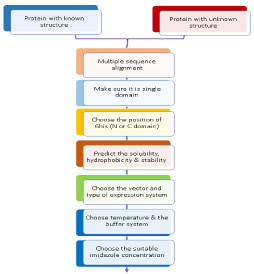
It is a well-known fact that the full-length proteins are difficult to be expressed in E. coli and mostly they get aggregated. Therefore, it is easy to crystallize the proteins containing single domain as compared to multi-domain proteins. The molecular weight of proteins also determines the success rate of crystallization. Moreover, the oligomeric state also favors the easy crystallization. Different strategies have been developed to achieve the higher rate of protein purification and crystallization. To enlist, different parameters like Isoelectric point, pH, domain organization, peptide signal, stability, prediction of secondary structure, a hydrophobic composition of protein, and salt concentration are fixed to achieve the goal of increased crystallization.
X-Ray crystallography is currently a powerful technique and has provided 3D structures of thousands of proteins. However, crystallization is still a largely empirical process. The production of the protein crystals is a prerequisite for such studies, which remains a rate-limiting step and the least understood. The principle of crystallization, both macromolecules and salts take a solution to the reservoir in high concentration and induce an exit solution; if this happens too fast then precipitation will occur, but under the right conditions it will increase crystals [4]. However, despite the growing advances, there are still many factors which limit the successful protein crystallization. Some known parameters like molecular weight, theoretical isoelectric point, the composition of amino acids, extinction coefficient help to increase the rate of successful protein purification. From the homologous protein structure, some variables like pH and salt concentrations are expected. The protein purity and homogeneity are crucial for successful protein crystallization. An initial screening is used for the protein crystallization by hit and trial method to find the right crystal condition. To favor the crystallization process, the solubility conditions of proteins in relation to favorable intermolecular interactions are altered. Moreover, it is important to try and rationalize the output of every condition; even the negative results will provide fruitful information and all the screen trials should be reviewed very carefully [5]. Further optimization is carried out by using various strategies to improve the shape and size of a crystal with varying pH, the concentration of salt, and the concentration of precipitant. Some protein crystals grow bigger in size by only varying precipitant which reveals that each component must be noted and have a great impact on the quality of crystal growth. Further additives screening is widely used for the new crystals which help to get different shapes of crystals and improve the diffraction of the protein crystal.
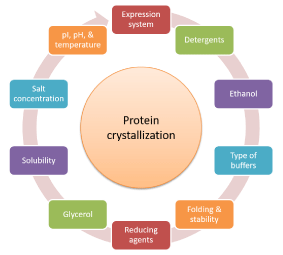
The crystallization process is driven by both thermodynamically and kinetically in which the solution reaches the supersaturation state. Protein crystals have been the subject of intense investigation for many years. Formation of crystal nuclei from supersaturated solutions does not specify that it will favor the large macroscopic crystals. It is a well-known fact that by crystallization, protein favors the protein stabilization. Proteins obtained in crystal form have the lower free energy of 3—6 kcal/mol than a dissolved molecule in a solution [5]. Typically, protein crystals are affected by the physical barriers which directly affect the protein crystallization like solution solubility, protein impurity, supersaturation, shape, size, the growth of the crystals, temperature, pH, nucleation, and buffer composition. In this article, some of the key factors are discussed that serves the base for achieving the better purification and crystallization (Figure 1).
Protein solubility
The protein chemists work hard to increase the protein solubility, although a challenging task. Low protein solubility is a factor in several types of diseases. To increase the efficiency of resolution for structural studies, different approaches have been followed to improve the protein, and solubility is of great interest to the structural studies [6,7], crystallization of membrane proteins [7,8,9], pharmaceutical applications [10,11], and treatment of human diseases [12,13,14]. The structural biologists work with soluble proteins is of extreme importance [7], pharmaceutical industry [15]. Some of the factors that make a big difference in protein solubility include temperature, pH, ionic strength, the rate of protein synthesis composition of protein, osmotic pressure, protein concentration, salt concentration and buffer composition and altering these factors lead to the successful solubility [7,15,16]. However, it is not appropriate for the protein solubility by varying the solution conditions. The amino acid composition on the surface of the protein influences the protein solubility. The solubility of proteins has shown an increase by the charged amino acids in a pH- and context-dependent manner [17,18]. Nevertheless, it is not well understood how one can alter the intrinsic properties of a protein to increase its solubility [7,16]. Recombinant protein purified by affinity chromatography consists affinity Tag (His/GST), which allows the protein to purify; it has been found that the biological activity is also affected by affinity tag. Conversely, there is no consolidated affinity tag differentiated as the best purification. In expressing a protein domain, the choice of affinity tag at N- or C- terminal gets affected, because a small difference influences the protein solubility. For example, a considerable variation in both solubility and aggregation operations by altering just a few amino acids in a protein length was examined on a nested set of 2,143 N- and C-terminal truncations from 96 targets by Klock and aolleagues18 [19]. It is wise to check which end of the protein is buried inside the fold. Moreover, if the three-dimensional structure is known it is recommended to keep the tag in solvent accessible- end. Some common examples of small peptide tags are the poly-Arg-, FLAG-, poly-His-, c-Myc-, S-, and Strep II- tags [20]. However, altering the Affinity tag from C-terminal to N-terminal or vice-versa of a protein has been found effective which showed to improve the solubility to some extent in many cases. The high concentration of proteins is required for various applications; protein crystallization is one among them and requires a desired concentration; however, a high concentration of protein often causes precipitation and aggregation. The addition of charged amino acids affects the protein solubility. The maximum protein concentration of a soluble protein can be achieved up to 8.7 times by adding the charged amino acids like L-Arg and L-Glu at a concentration of 50mM to the buffer and prevents protein precipitation and aggregation over time gives long-term stability and prevents protein-protein and protein- RNA interaction [21].
Imidazole effects in protein solubility and labeling reactions
Imidazole is extensively used in protein purification processes especially in protein elution using nickel column. A high concentrated imidazole (1M) containing protein sample should be removed by using the additional purification steps following the nickel affinity column. To remove the imidazole, size exclusion chromatography (SEC) is an appropriate and potential technique. Moreover, imidazole concentration can be simply reduced from 1M to a reasonable level (0.02 – 0.2 M) including for crystallization reagents if the imidazole is inconsistent with the sample homogeneity. In addition, it has been seen that imidazole might affect the protein crystallization and one can try to remove the imidazole by precipitating the protein in a solution of ammonium sulfate and keep back in low salt concentration buffer followed the purification by ion exchange or SEC. Dialysis is another good way to remove the imidazole from the protein sample. In one instance, by using 0.6 M imidazole/acetate buffer sufficient crystals could only be obtained and were seen protein surface containing hydrophobic spots occupied by two molecules of imidazole. In some cases, it has been seen that purified protein from affinity Ni column precipitate without imidazole might be the reason that Ni ions leak from Ni- column. In addition, his-Tagged protein aggregates in solution upon the removal of Ni, as the presence of imidazole may have been chelating with this excess and trace nickel. To overcome this problem, one can use Ni-chelating resin to remove this excess nickel. Furthermore, a reasonable concentration (0.02 – 0.1 M) of imidazole can be used with the sample or EDTA or citrate buffer. Researchers working with metalloprotein and need metals for the protein stability might use the chelating resin instead of leaving a chelating reagent in the sample as it is apparently a better choice
Effect of protein precipitation during dialysis
It is essential to examine the influence of protein concentrations on the precipitation of target protein after storage. Generally, proteins stored with concentration are much better than diluted one. Protein sample with 10 mg/ml will have relatively less loss of adsorption than at low concentration 0.5 mg/ml. However, the too concentrated sample also leads to precipitation, it is recommended to store the protein at dilute concentration and concentrate the sample right prior to crystallization.
Expression system
The bacteria are the most widely used microorganism specifically Escherichia coli to produce the recombinant proteins. It is the first option for structural biologists to produce high-level recombinant protein for x-ray crystallographic studies and in vitro biochemical assays, bacteria are easy to modify genetically and produce recombinant proteins. Plenty of designed expression systems are available for biological and commercial applications and the three-dimensional protein structures are solved and submitted in Protein Data Bank (PDB) in 2003 which determines 90% E. coli expression system, moreover pET expression system based T7 promoter is most widely used expression system and contributes 90% in 2003’s PDB protein preparation systems [22]. The most widely used common promoters include Trc promoter (e.g., Amersham Tac promoter (e.g., Amersham-PL promoter/cI repressor (e.g., Invitrogen pLEX), Biosciences pGEX) Biosciences pTrc) and hybrid lac/T5 (e.g., Qiagen pQE) Promotors [22]. It is very fast and inexpensive and can be used to test a wide spectrum of proteins to make a comprehensive analysis within a short period of time. Over a past few years, much effort has been done for optimizing bacteria (E. coli) as a host expression system and a wide arsenal of tools has been generated by this strategy to increase the yield of protein solubility [23]. A BL21 (DE3) is an E. coli strain, deficient in both Ion and Omp proteases and compatible with the T7 lacO promoter system, used for high-level protein production [24]. Bacteria do not require post-translational modifications and produce only small proteins. Being prokaryotic, limited proteins can be expressed because it does not perform the post-translational modification, folding machinery, chaperons which limit its use for the expression system. It is often difficult to express in terms of full-length protein in E. coli, up to 50% proteins from Eubacteria or Archaea and 10% of proteins from eukaryotic can be expressed in E. coli in soluble form as shown by the large-scale protein expression trials [25]. High molecular weight proteins have been expressed in the pellet (insoluble aggregates), known as inclusion bodies which are cytoplasmic, insoluble aggregates of protein. However, there are the developed methods for the processing of these inclusion bodies which make them soluble by using standard concentration of denaturing agents. The denaturing agents that are used include Guanidine.HCl, Urea, Guanidine thiocyanate, Sodium dodecyl sulfate (SDS), n- lauroyl sarcosinate (sarkosy). Among them Guanidine.Hcl and Urea are the most common denaturing agents for protein denaturation and then renaturalize the protein to its active form. In the solution, the hydrophobic surfaces, especially with planer amino acid side chains (Arg, Trp Gln) and aliphatic side chains, are coated by these guanidium ions which reduce the unfavorable exposure from the solvent. The exposed peptide groups in the protein are surrounded by the urea rather appear as a stacking process. The major contribution to the protein stability is the hydrophobic effect, therefore, weakening by the guanidine and urea [26]. However, this process is time-consuming and yield is not much. In some cases, expression level has been seen very low after induction because of the toxicity of protein which not only inhibits the cell growth but also kills the cells. The E. coli may not express longer protein > 80 kDa; there, it is suggested to shorten the length of a protein fragment, delete or mutate the hydrophobic residues which increase the protein expression and solubility. There are other choices for improving expression and purification levels of different proteins by using Yeasts, Insect cell-based expression systems, Mammal cells as production factories, transgenic animals, and transgenic plants which express full-length proteins, longer fragments, although it takes several weeks to months to express the protein.
Folding and stability
The stability of the proteins in various buffer compositions and different pH values with ligands or without ligands can be characterized whether the protein is correctly folded by using the CD (Circular dichroism) spectroscopy, UV/Visible Spectrophotometry, NMR and differential scanning fluorimetry (DSF). Useful websites for fold recognition can be used to predict the protein fold (RF-Fold, SSHMM, THREADER, BLASTLINK, SSEARCH, PSI-BLAST and HMMER).
Some proteins are not correctly folded and require cofactor, prosthetic group or ligand to make the protein properly folding and hence, increase the stability. It is more likely that beta-sheets form amyloid-like aggregates that can be crystallized in presence of other binding partners, which helps in stabilization and protein folding. A misfolded or unfolded protein is the disruption of native state and is a nonfunctional protein which is thermodynamically unstable tending to form aggregates. Disulfide bonds are believed to increase the stability of the protein by decreasing the conformational entropy of the unfolded state [27]. Alpha helices are stabilized by predominantly hydrogen bonds, while hydrogen bonding and hydrophobic interactions are the stabilizing forces for beta sheets. Exposed hydrophobic residues in a protein molecule in the cell due to misfolding or mutation tend to stick together and lead to the formation of insoluble aggregates called inclusion bodies in recombinant proteins (Figure 2).
Reducing agents
The cysteine-containing proteins often become a problem in purification due to oxidation, which causes aggregation. To prevent the oxidative cross-linking of thiol groups, sulfhydryl reagents (reducing agents) such as DTT, B-ME, and TCEP are commonly added to the buffer. The TCEP is the most stable among them; however, expensive and at neutral pH it is not stable, particularly in phosphate buffers. The aim of adding reducing agents is to reduce the aggregation which helps the crystallization process. However, reducing agents can interact with the metal proteins which affect the crystallization. Moreover, DTT is more advantageous than B-ME because of shorter half-life, but having low volatility. Variation of pH affects both, BME has a half-life greater than 100 hours and DTT has 40 hours respectively at pH 6.5 (20ºC). At pH 8.5 (20ºC) half-lives reduce to 4 h and 1.4 h respectively [28]. Dithiothreitol (DTT), also known as Cleland’s reagent, is used for protein reduction. Dithioerythritol (DTE), an isomer having similar properties, has been used in reduction, except that the former is more effective. These buffers should be stored at 4 degrees and add reducing agents when you are ready to use. BME reacts easily with cobalt, copper and other phosphate buffers while DTT reacts easily with nickel. It is also important not to use it in high concentration; it affects the column and reduces the nickel in the nickel column which turns brown. Nevertheless, it is not reported yet which reducing agent is best to be used for successful protein crystallization.
Isoelectric point (pI) and pH
Every protein has its isoelectric point at which the net charge of the protein is zero. At that point, protein does not move and possibly it aggregates. Acidic proteins having isoelectric point lower than 7 are more likely to crystallize one pH unit above their isoelectric point, while basic proteins having lower than their isoelectric point are likely to crystallize at about 1.5–3 pH units, the stability, and solubility of the protein is affected by different pH values. The pH of the buffer component is considered to be the most significant parameter rather than pH of the final crystallization solution, which can vary up to three pH units [29]. In general, protein soluble at lower pH (Protonate), protein soluble at high pH (deprotonate). To move the pH away from pI increases the solubility of the protein, and more likely to decrease the electro-repulsion at low salt concentrations and prevent the aggregation and precipitation problems. Moreover, ligand binding increases the stability of the protein structure, mostly in case of enzymes. If there is an increased difference between pH and pI, higher salt concentrations are required for crystal growth.
Temperature
Temperature can be a remarkable variable in crystallization of biological macromolecules (Proteins) [30]. The nucleation and crystal growth is often affected by temperature by influencing the solubility and super-saturation of the sample. Temperature is one of the most important factors affecting both protein purification and crystallization steps. Most proteins are functional at a lower temperature and are purified at 4ᵒC or in cold room. However, some proteins are stable and purified at room temperature. Usually, proteins use crystallization screening and are performed at 20ᵒC and sometimes 4ᵒC. Proteins screened and optimized at a reasonable temperature range between 4-45 ᵒC, although some proteins have been crystallized at 60ᵒC (glucagon and choriomammotropin). Temperature influences the protein crystallization by decreasing the solubility, crystal growth, packing, termination and on crystal nuclei formation; a small change in temperature makes a big difference in the crystallization process. Purified proteins with high salt concentration and with normal solubility will be soluble at a cold temperature than the room temperature, while protein with normal solubility in low salt will be more soluble at warm than at cold temperatures. Temperature affects the pH of most buffers, especially the tris buffers. The precipitate or crystal form in a lower concentration of PEG, MPD, or organic solvent by proteins with ‘normal’ solubility is slower at lower than at higher temperatures.
Salt
Finding the right buffer to keep a protein in solution and obtain optimal activity is a trial-and-error process. In general, starting from 50mM-150 mM NaCl (Normal) concentrations is a good starting point. In SEC, usually, 150 mM NaCl (Normal) concentration is used to keep proteins in soluble form and mimics their physiological conditions for the successful protein purification. Different concentration of salt with different pH values can be used for better purification and crystallization. If proteins do not crystallize at normal buffer (150mM NaCl), must be purified with high salt concentration (1M NaCl) at different pH values which helps to reduce the oligomerization and aggregation. The aim of high salt concentration is luckily found fruitful for successful crystallization because it stabilizes the protein buffer. A high concentration decreases the solubility which leads to precipitation. Its effect can be seen on crystallization success that happens in the drop where all components get concentrated through the water loss. If the concentration of protein is too low, it will not reach its marginal value and the drop will be seen very clear. In such cases, the concentration of the protein should increase. The use of 50mM to 1M NaCl concentration can be used to make successful crystallization. However, it is important to keep the concentration of salt in a buffer optimized for the successful crystallization process. In case of other chromatography techniques, for example, ion exchange, one should start at a low salt concentration (5-25 mM) for protein purification which helps to prevent nonspecific binding and ionic interactions to the column.
Size exclusion chromatography (SEC)
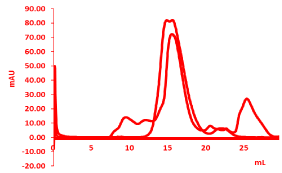
Size exclusion chromatography (SEC), used for the protein purification, provide a successful protein purification based on the size and molecular weight of the molecule. The purity of target protein first should be verified from SDS –PAGE, and then used for size-exclusion chromatography (SEC); Superdex 200 size-exclusion column 10/30 (GE Healthcare, Princeton, NJ, USA). Ion exchange chromatography can be used further to increase the purity. The purity and homogeneity are crucial for the successful protein crystallization as shown by the two different peaks for good and bad proteins shown in figure 3 of gel filtration peaks. An oligomeric form of the protein will affect the crystallization and will be hard to crystallize, further characterization can be done by using the MALS (Multi-angle light scattering) which gives the stoichiometry of the molecule. It will give the exact molecular weight and subunit constituents with polydispersity.
Glycerol
Glycerol is a polyalcohol, widely used to maintain the protein in solution by increasing the solubility. The protein structure is stabilized by glycerol [30]. So, glycerol is often used in crystallization solvent, moreover, if crystallization occurs over a longer time, it is used mainly for this purpose. Along with increasing glycerol concentration, there is also the need of increasing the larger amounts of salt concentration to crystallize the protein. It acts as a cryoprotectant, used before mounting of crystals because of having antifreeze property. The effect of glycerol on protein-protein interaction has been reported in the previous studies. Farnum and Zukowski have shown that there is an increased protein-protein repulsion in the aqueous solution of bovine pancreatic trypsin inhibitor on the addition of glycerol [31]. It has been reported that crystallization takes place at a fixed protein concentration when the amount of salt is increased. Because the increasing glycerol concentration progressively takes a long time for crystallization. Glycerol works as an anti-nucleation agent at some time in crystallization.
Mutation
Proteins contain several distinct domains that are tethered by the flexible linkers making a modular like structure. When the protein solubility is limited and failure in crystallization after using well-defined crystallization screening conditions, it is better to work with truncated, point mutated, single domains or subunits of multimeric proteins rather than the native proteins. Changing a single amino acid has shown dramatic changes in the protein crystallization properties, this could be due to change in the charge or polarity of the wild-type amino acids; however, there is no correlation between the solubility trends and crystallizability, yet, its effect has been seen on crystallization step. The engineered crystals are produced by making the point mutations, thereby generating a number of engineered crystals that include human H-chain ferritin, human thymidylate synthase, E. coli glutathione reductase, human leptin, the horse spleen ferritin and rat liver L-chain ferritin. The Gln is absent in human H-Chain instead of Lys86 by replacing the Lys86 to Gln in human H-chain resulting in the formation of the crystal, diffracted to 1.9A0 resolution. However, it is still not understood which specific mutation leads to successful crystallization.
Post-translational modifications
Post-translational modifications (phosphorylation, glycosylation, and ubiquitination) have significant effects on the solubility of proteins produced. The post-translational modification can be acquired at any step in the proteins life cycle. It is a well-known fact that disordered proteins tend to be phosphorylated at flexible regions which result in disorder-to-order as well as order-to-disorder transitions [32]. In addition, Physico-chemical properties, stability, kinetics, and dynamics can be altered by adding or removing dianionic phosphate group somewhere on the protein. Near Binding interface where the phosphorylation occurs, which influences the binding energy of the complex. Moreover, phosphorylation takes place at a site outside the binding interface may cause the conformational changes through an allosteric mechanism which causes to the effect of binding partner [33]. Phosphorylation affects the complex stability and sometimes it was supposed to be mutant phosphorylated sites (Serine, threonine, and tyrosine) especially those on the protein surface if it is eukaryotes protein and Histidine in prokaryotes and plants. Nevertheless, it is important to keep in mind that phosphorylation acts for the degradation of some protein. If the protein contains multiple sites, they are phosphorylated, therefore, it is advised to reduce the number of the site by mutating it.
Conclusive remarks
In this review, we have focused on the successful key factors which affect the protein purification and crystallization processes. We have explained the possible ways to obtain high-level protein purification, as well as, high-quality protein crystals, and provide the ways to improve the results. Moreover, we explained how structural state of protein could be affected by critical parameters such as metal ions, cofactors, inhibitors, Iso electric point, temperature, SEC, glycerol pH, salt, buffer, post translational medications etc. The critical parameters specifically purity and homogeneity have been critically examined for successful protein crystallization. A negative effect could be observed in identifying the right crystallization condition if compromise can be made either of them. Furthermore, an attempt has been done for reporting the other challenges for successful protein purification and crystallization.
- Lorber B, Fischer F, Bailly M, Roy H, Kern D (2012) Protein analysis by dynamic light scattering: methods and techniques for students, Biochem Mol Biol Educ 40: 372-382. Link: https://tinyurl.com/y8rkqsvv
- Rudolph R, Lilie H (1996) In vitro folding of inclusion body proteins, FASEB J 10: 49-56. Link: https://tinyurl.com/yae7fkre
- Fischer B, Perry B, Sumner I, Goodenough P (1992) A novel sequential procedure to enhance the renaturation of recombinant protein from Escherichia coli inclusion bodies, Protein Eng 5: 593-596. Link: https://tinyurl.com/y9mypobf
- Gernert KM, Smith R, Carter DC (1988) A simple apparatus for controlling nucleation and size in protein crystal growth, Anal Biochem 168: 141-147. Link: https://tinyurl.com/y8xly7qc
- Benvenuti M, Mangani S (2007) Crystallization of soluble proteins in vapor diffusion for x-ray crystallography, Nat Protoc 2: 1633-1651. Link: https://tinyurl.com/y9jcrsvd
- Ries-Kautt M, Ducruix A (1997) [3] Inferences are drawn from physicochemical studies of crystallogenesis and crystalline state, Methods Enzymol 276: 23-59. Link: https://tinyurl.com/y8kjw8tn
- Bagby S, Tong KI, Ikura M (2001) Optimization of protein solubility and stability for protein nuclear magnetic resonance, Methods Enzymol 339: 20-41. Link: https://tinyurl.com/ydbrtg65
- Frank S, Kammerer RA, Hellstern S, Pegoraro S, Stetefeld J, et al. (2000) Toward a high-resolution structure of phospholamban: design of soluble transmembrane domain mutants, Biohemistry 39: 6825-6831. Link: https://tinyurl.com/yczj5jbn
- Slovic Am, Kono H, Lear JD, Saven JG, DeGrado WF (2004) Computational design of water-soluble analogs of the potassium channel KcsA, Proc Natl Acad Sci USA 101: 1828-1833. Link: https://tinyurl.com/y79x86kp
- Fowler SB, Poon S, Muff R, Chiti F, Dobson CM, et al. (2005) Rational design of aggregation-resistant bioactive peptides: reengineering human calcitonin, Proc Natl Acad Sci U S A 102: 10105-10110. Link: https://tinyurl.com/yc7c92lc
- Ricci MS1, Brems DN (2004) Common structural stability properties of 4-helical bundle cytokines: possible physiological and pharmaceutical consequences, Curr Pharm Des 10: 3901-3911. Link: https://tinyurl.com/yasjqblo
- Brouet JC, Clauvel JP, Danon F, Klein M, Seligmann M (1974) Biologic and clinical significance of cryoglobulins. A report of 86 cases, Am J Med 57: 775-788. Link: https://tinyurl.com/ybml3jr4
- Evans P, Wyatt K, Wistow GJ, Bateman OA, Wallace BA, (2004) The P23T cataract mutation causes loss of solubility of folded gammaD-crystallin, J Mol Biol 343: 435-444. Link: https://tinyurl.com/ybcuqqld
- Pande A, Annunziata O, Asherie N, Ogun O, Benedek GB, et al. (2005) Decrease in protein solubility and cataract formation caused by the Pro23 to Thr mutation in human gamma D-crystallin, Biochemistry 44: 2491-2500. Link: https://tinyurl.com/y9suc3af
- Caldwell GW, Ritchie DM, Masucci JA, Hageman W, Yan Z (2001) The new pre-preclinical paradigm: compound optimization in early and late phase drug discovery, Curr Top Med Chem 1: 353-366. Link: https://tinyurl.com/ydhvq4oo
- Schein CH (1993) Solubility and secretability, Curr Opin Biotechnol 4: 456-461. Link: https://tinyurl.com/y8huhhrn
- Trevino SR, Scholtz JM, Pace CN (2007) Amino acid contribution to protein solubility: Asp, Glu, and Ser contribute more favorably than the other hydrophilic amino acids in RNase Sa, J Mol Biol 366: 449-460. Link: https://tinyurl.com/y9z28vy6
- Islam MM, Khan MA, Kuroda Y (2012) Analysis of amino acid contributions to protein solubility using short peptide tags fused to a simplified BPTI variant, Biochim Biophys Acta 1824: 1144-1150. Link: https://tinyurl.com/y8wf6n2l
- Graslund S, Nordlund P, Weigelt J, Hallberg BW, Bray J, rt al. (2008) Protein production and purification, Nat Methods 5: 135-146. Link: https://tinyurl.com/ya2ngpal
- Terpe K (2003) Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems, Appl Microbiol Biotechnol 60: 523-533. Link: https://tinyurl.com/yb8nltfx
- Golovanov Ap, Hautbergue GM, Wilson SA, Lian LY (2004) A simple method for improving protein solubility and long-term stability, J Am Chem Soc 126: 8933-8939. Link: https://tinyurl.com/yb25frc3
- Sørensen HP1, Mortensen KK (2005) Advanced genetic strategies for recombinant protein expression in Escherichia coli, J Biotechnol 115: 113-128. Link: https://tinyurl.com/ydgy7vpe
- Peti W, Page R (2007) Strategies to maximize heterologous protein expression in Escherichia coli with minimal cost, Protein Expr Purif 51: 1-10. Link: https://tinyurl.com/y8urgfdu
- Studier FW, Rosenberg AX, Dunn JJ, Dubendorff JW (1990) Use of T7 RNA polymerase to direct expression of cloned genes, Methods Enzymol 185: 60-89. Link: https://tinyurl.com/y7y8uea5
- Braun P, LaBaer J (2003) High throughput protein production for functional proteomics, Drug Discov Today 9: 1-7. Link: https://tinyurl.com/y9yg69gz
- England JL, Haran G (2011) Role of solvation effects in protein denaturation: from thermodynamics to single molecules and back, Annu Rev Phys Chem 62: 257-277. Link: https://tinyurl.com/ya3t6ljj
- Zavodszky M, Chen CW, Huang JK, Zolkiewski M, Wen L, et al. (2001) Disulfide bond effects on protein stability: designed variants of Cucurbita maxima trypsin inhibitor-V, Protein Sci 10: 149-160. Link: https://tinyurl.com/ybmypm3n
- Abdalla M, Ali Eltayb W, Samad A, Elshareef SHM, Dafaalla TIM (2016) Important Factors Influencing Protein Crystallization. Glob J Biotechnol Biomater Sci 2: 25-28. Link: https://tinyurl.com/yb6hcual
- Kirkwood J, Hargreaves D, O'Keefe O, Wilson J (2015) Analysis of crystallization data in the Protein Data Bank, Acta Crystallogr F Struct Biol Commun 71: 1228-1234. Link: https://tinyurl.com/y7a87pc6
- Junius N, Oksanen E, Terrien M, Berzin C, Ferrer JL, et al. (2016) A crystallization apparatus for temperature-controlled flow-cell dialysis with real-time visualization, J Appl Crystallogr 49: 806-813. Link: https://tinyurl.com/ycsszpws
- Farnum M, Zukoski C (1999) Effect of glycerol on the interactions and solubility of bovine pancreatic trypsin inhibitor, Biophys J 76 2716-2726. Link: https://tinyurl.com/y8au74np
- Antz C, Bauer T, Kalbacher H, Frank R, Covarrubias M, et al. (1999) Control of K+ channel gating by protein phosphorylation: structural switches of the inactivation gate. Nat Struct Biol 6: 146–150. Link: https://tinyurl.com/y83awnp6
- Nishi H, Fong JH, Chang C, Teichmann SA, Panchenko AR (2013) Regulation of protein-protein binding by coupling between phosphorylation and intrinsic disorder: analysis of human protein complexes Mol Biosyst 9: 1620-1626. Link: https://tinyurl.com/ycobbwyg
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
