Forensic Science Today
Long-term persistence of Saliva-derived microRNA in human dental calculus for Forensic investigations
Catia Lippolis1*, Aldo Cavallini1, Ginevra Panzarino2 and Sandro Sublimi Saponetti2
2Laboratory of Anthropology, Department of Biology, “Aldo Moro” University of Bari, via Orabona 4, Bari 70125, Italy
Cite this as
Lippolis C, Cavallini A, Panzarino G, Sandro Saponetti S (2019) Long-term persistence of Saliva-derived microRNA in human dental calculus for Forensic investigations. Forensic Science Today 5(1): 001-007 DOI: 10.17352/fst.000012Introduction: Human identification is one of the major fields in forensic odontology especially when the dental remains are the only available evidence to individual identification. In this context DNA analysis in teeth pulp remains the most accurate and reliable method for identification, whereas RNA is less used for forensic purpose because it is generally considered a quickly degradable molecule. Recently some studies report that another excellent source of nucleic acids is the dental calculus or tartar.
Method: Therefore to assess the RNA stability over time, we investigated 40 saliva-derived microRNAs (miRNAs), in archeological and modern tartar samples by classic PCR followed by quantitative PCR.
Result: Three miRNAs were not present in both ancient and modern tartar samples, while 10 out of 37 miRNAs had significant different levels in ancient than modern samples. Nine miRNAs were down-regulated and only one was up-regulated. Since these different levels were not induced by gum inflammation and were independent to guanine-cytosine content in miRNA sequences, used as degradation biomarker, we hypothesized as a possible cause the different eating habits between ancient and modern populations. This hypothesis is also supported by recent studies that demonstrate an close relationship between nutrients and changes in miRNA expression in healthy individuals.
Conclusion: Overall data show that miRNAs are more stable than other RNA types in ancient tartar and this could enlarge the possibility of their practical use for forensic purpose and bioarchaeological researches although to better understand the variability of some miRNA levels between ancient and modern populations requires further investigation.
Abbreviations
BC: Before Christ; AD: Anno Domini; RT: Reverse Transcriptase; qPCR: quantitative Polymerase Chain Reaction; preAmpPCR: pre-Amplification PCR; LNA: Locked Nucleic Acid; rRNA: ribosomal RNA; miRNA: microRNA; IQR: Inter-Qrange; GC: Content: Guanine-Cytosine Content.
Introduction
Forensic odontology, a branch of Forensic sciences, is important in the identification of the dead person. In the last years this scientific discipline has assumed an increasingly important role in personal identification also supported by recent advances in the knowledge and techniques of molecular biology [1]. Generally the forensic odontology examines injuries to jaws, teeth, and oral soft tissues, however in cases of body carbonization, blast injuries or mass disasters situations [2], the teeth, the most indestructible components of the human body, could be the only mean of identification of an otherwise unrecognizable body [3,4]. It is also known that dental pulp and dentin are better sources of DNA than skeleton bones and although DNA undergoes progressive fragmentation through autolytic and bacterial enzymes, the sequence of information is still present in its fragments [5]. Another excellent source of nucleic acids is the dental calculus, also known as tartar, that shows more abundant and less contaminated DNA as compared to that extracted from dentin [6]. Several studies have investigated human and bacterial DNA in archaeological dental calculus but few systematic studies have been performed on RNA because this molecule is generally considered fragile and it is readily degraded than DNA in body decomposition [7]. However a recent archeological study has identified and analyzed microRNAs (miRNAs), small non-coding RNAs, in human specimens thus showing that these small molecules could survive long-term post-mortem [8].
miRNAs are small RNA molecules of 20-23 nucleotides long that are involved in many biological functions and their abnormal levels in biological fluids, including saliva, have been used as possible biomarkers in various human diseases, including tumors [9].
The molecules present in tartar matrix are those contained in the saliva and in order to investigate miRNA stability in this biological matrix during a long period of conservation under unfavorable conditions like burial we have analyzed and compared miRNA expression in archeological and modern tartar samples. Forty saliva-derived miRNAs found highly expressed in healthy subjects were chosen from literature data and analyzed in our study [10-12].
Since that miRNAs represent a minor fraction of the total RNA [13] and the success of RNA analysis is also affected by the laboratory methods with which it is processed, we expected to find very low amounts of miRNAs in the tartar samples and therefore the use of a highly sensitive detection technique was needed. Comparing 12 different commercially methods for miRNA analysis, Mestdagh et al. have indicated that the sensitivity was very much technology-related qPCR especially when a sample contained low miRNA levels [14]. In addition, the RT-qPCR method was also used in a previous study to analyzed miRNA profiling in highly degraded RNA extracted from archeological specimens [10]. Therefore we have applied in our study a sensitive PCR-based method consisting of two phases: a pre-amplified reverse-transcribed cDNA (RT-preAmpPCR) followed by quantitative real-time PCR (qPCR) assay.
Materials and Methods
Modern and ancient dental calculus manipulation and RNA extraction
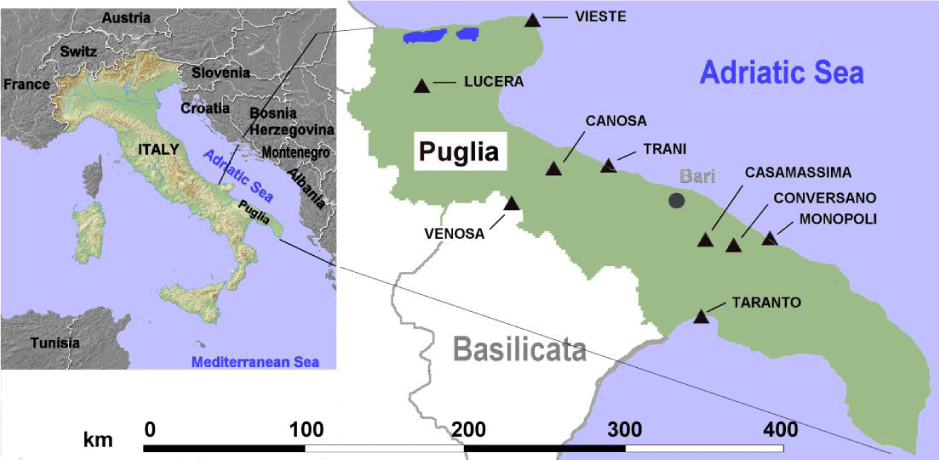
Archeological tartar samples were obtained from 20 individuals recovered from 11 different Puglia burial sites (Figure 1). All individuals covered a period extending from 9th century BC to 15th century AD. We selected human remains buried in graves rather than in the soil because the state of skeleton preservation was very high. In addition, anthropological evaluation did not show signs of bone alterations due to diseases. Sex, age and dating of the time of death were evaluated by standard anthropological instruments and morphological characteristics were performed according to methods of Beck [15]. The age of individuals was of 39.7 ± 15.3 years (mean ± SD), 15 were males and 5 females. List and characteristics of selected human remains are provided in table 1.
Modern tartar samples were obtained from 30 healthy volunteers undergoing a dental examination at a private clinic of oral medicine and periodontology of Bari (Puglia, Italy). This group was composed of 16 females and 14 males with an average age of 49.1 ± 12.3 years (mean ± SD). All subjects reported no systemic diseases and dental specialists observed no signs of oral diseases.
All tartar samples were removed in sterile conditions by curette and placed in sterile tube. The pulverization of the samples was carried out in a clean room facility of Department of Anthropology, University of Bari and to avoid contamination standard criteria described by Kirsanow and Burger were observed [16]. In brief, the tartar samples were subjected to UV irradiation (254 nm) to eliminate surface contamination, submerged in a sterile container (petri dish) containing molecular grade bleach (4% final dilution) for 5 min and then rinsed with ethanol (90%) for 3 min to remove traces of bleach. Afterwards the samples were pulverized in liquid nitrogen by mortar and pestle and the powder was transferred to a sterile tube avoiding cross-contamination between ancient and modern samples. In order to monitor a possible external DNA/RNA contamination, a series of control tubes contained DNAse/RNAse free water instead of tartar powder were included during sample collection and manipulation.
The permission to remove ancient tartar from human remains was obtained from Sovrintendenza dei Beni Culturali of Puglia (Italy), while modern tartar samples were extracted from teeth of healthy volunteers after obtaining informed consent.
Tubes with tartar powder, as well as control tubes, were transported to Molecular Biology lab of Castellana Grotte (BA) for miRNA analysis. Since a PCR laboratory workflow should be designed to prevent external contaminations, such as qPCR products and cross-contamination between samples [17], the setting up of a suitable qPCR-facility is always needed to perform forensic procedure or medical diagnostic assays. Therefore to minimize the risk of contamination, the tartar RNA extraction and next amplification were set up following general guidelines previously described [18].
After UV radiation overnight, 30 mg of powder were subjected to RNA extraction with phenol-chloroform solution (TRIzol reagent, Thermo Fisher Scientific, Rodano (MI), Italy), followed by use of miRNeasy mini kit (Qiagen, Milan, Italy) to eliminate any PCR inhibitors present in tartar matrix. All procedures of RNA extraction were performed according to manufacturers’ instructions. Total RNA was quantified by two different methods, spectrophotometric (A260/280) and fluorimetric (Qubit device, Thermo Scientific) methods to obtain an accurate value of total RNA. The total RNA was expressed as ng/30mg of tartar powder. In order to evaluate human RNA in total RNA, the bacterial RNA was isolated by MICROBEnrich kit (Thermo Fisher Scientific). The MICROBEnrich kit is based on hybridization of poly (A)-tails of human 18S and 28S rRNA that constitute 80–90% of total cellular RNAs. However, ancient human RNA is commonly fragmented and part of 18S and 28S rRNA may not be extracted by MICROBEnrich kit for the loss of the poly (A)-end and be present in eluate. Therefore, in order to evaluate the loss of human RNA by using this kit, the rRNA levels were analyzed by 18S and 28S rRNA Control kits (Eurogentec, Seraing, Belgium) in both retained and eluted fractions. To detect the rRNA levels, two PCR steps: RT-preAmpPCR followed by qPCR step. This PCR method is explained later in miRNA evaluation. To normalize rRNA expression profiling, Copy number standard curve kit (Primerdesign Ltd, Chandler’s Ford, UK) was used following the manufacturer’s instructions. Finally, total human RNA was expressed as pg/ng of total RNA.
PCR method and miRNA evaluation
The miRNA levels were evaluated by a RT-PCR method. After reverse-transcription, cDNA was amplified by two successive PCR runs, also called nested-PCR, to increase the method sensitivity. The nested-PCR was composed of a classic preAmpPCR for 5 cycles followed by qPCR step for 40 cycles. Both PCR steps with minor changes in the procedures were described previously [19,20]. In brief, aliquots of total tartar RNA (30pg) were reverse-transcribed in 20 µl by miRCURY LNA RT Kit (Exiqon-Qiagen) in accordance to manufacturer’ instructions. Two out of 20µl was diluted with 158 µl of DNase and RNase-free water and 4µl were then pre-amplified in 50 µl for 5 cycles by preAmpPCR (classic PCR) and 5 µl of preAmpPCR product were then subjected to qPCR using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, Milan, Italy) according to manufacturer’s instructions. The same primers for each miRNA were used for both PCR steps. The primers were formed with Locked Nucleic Acid (LNA) (Exiqon-Qiagen) technology based on the use of chemically modified primers that allowed a high affinity hybridization between primers and miRNA target.
No-template controls (control tubes) were inserted between two PCR phases to exclude the cross-contamination risk.
To improve the PCR specificity, the annealing temperature during PCR assay was carried out at 65°C instead of 60°C as previously described [20]. To avoid false positive results in qPCR analysis, Cq values ≥ 35 cycles were arbitrarily discarded. Each qPCR was run in triplicate to ensure consistency and reproducibility of the results, while miRNA levels were evaluated with the following formula: log (2-ΔCq), where ΔCq = Cq miRNA target – Cq U6, where U6 snRNA, non-coding small nuclear RNA, was used as reference.
Since the amplification process consisted of two steps, a critical factor in the accuracy of the analysis could be a different PCR efficiency, expressed by the following formula: E = 10(-1/slope), between the two PCR phases.
To monitor PCR efficiency we used UniSp6 template (cat. n. YP00203954, Exiqon-Qiagen) in preliminary experiments. Serial dilutions of UniSp6 (10.0, 1.0, 0.01 and 0.001 fmol) were separately subjected to qPCR (35 cycles) and preAmpPCR (5 cycles) followed by qPCR (30 cycles), respectively. The Cq values were plotted against the log of the UniSp6 dilutions and linear regressions were performed from which the mean E was derived. The average value of E was 98% for qPCR and 96% for preAmpPCR followed by qPCR. In order to evaluate tartar miRNA recovery, the same UniSp6 concentrations described above were added separately to a fix tartar powder and, after extraction and RT phase, were subjected to preAmpPCR followed by qPCR.
To monitor miRNA degradation, we analyzed the guanine-cytosine (GC) content in miRNA sequences because it is known that miRNAs with low GC content are more degraded than GC-rich miRNAs [21], while 28S:18S rRNA ratio, another biomarker for RNA integrity, was excluded because some authors have shown that this ratio could be affected by cell aging [22]. The miRNA sequences are reported in table 2.
The microorganisms present in tartar matrix are contaminants that could alter the miRNA levels. However, a recent archaeological study did not find a relationship between bacterial genome and human miRNA sequences [10]. Furthermore a limited number of miRNAs or miRNA-like RNAs have been discovered in the microorganisms today [23] and the tartar is less vulnerable to contamination by non-human DNA than bone or dentine samples [24]. For all these reasons the bacteria were not considered as miRNA contaminants in our study.
Statistical analysis
The miRNA levels were expressed as median and interquartile range (IQR) and their differences between modern and ancient tartar samples were evaluated by non-parametric Mann-Whitney test. The correlation between miRNA levels, expressed as means, and their CG content was evaluated by Pearson’s correlation.
All statistical analyses were performed by GraphPad Prism software version 5.0 (GraphPad software, Inc., La Jolla, CA, U.S.A.) and p value < 0.05 was considered significant.
Results
RNA content in modern and ancient dental calculus
The total RNA amount recovered from tartar samples was significant different between modern and ancient samples with values of 4.6 ± 2.8 and 2.4 ± 1.6 ng RNA/30mg of tartar (means ± SD, p = 0.042), respectively.
Human 18S and 28S rRNA concentrations were 4.3 ± 3.0 and 1.1 ± 0.9 pg/ng total RNA (means ± SD, p = 0.031) in modern and ancient samples, respectively. In particular the 18S rRNA fragmentation find in eluate fractions ranged from 30.4% to 57.3% (median 45.4%) in ancient samples and from 20.7% to 38.1% (median 29.8%) in modern samples. Similar DNA loss were found by Kemp et al. in aged and degraded biological samples [25].
miRNA expression
A good recovery was obtained by using synthetic UniSp6. The recovery value was 82.5% ± 4.6% (mean ± SD) and 80.0% ± 5.7% (mean ± SD) in modern and ancient samples, respectively. All control tubes used to monitor the contamination showed no PCR products.
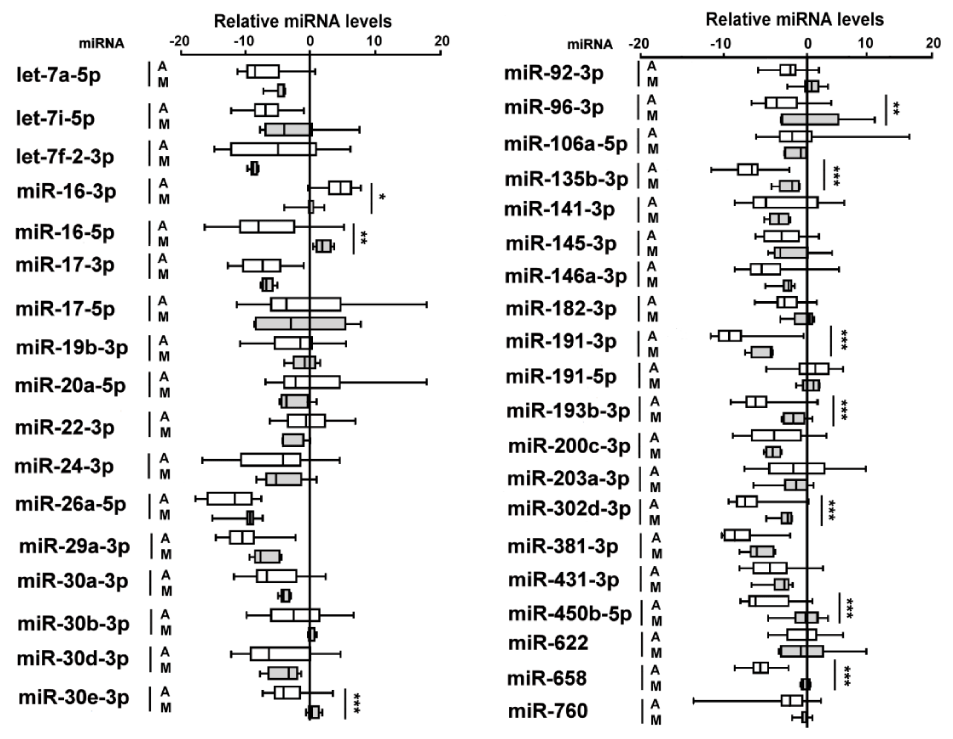
After amplification process miR-125a-5p, miR-195-5p and miR-212-5p were not detected in both sample groups. The same results were obtained when saliva samples of 10 out of 30 healthy volunteers, chosen at random, were analyzed to confirm the absence of three saliva-derived miRNAs in the tartar samples (data not shown) [26].
Thirty-seven miRNAs were present in all tartar samples and their levels were normalized by U6 RNA reference that did not shown appreciable fluctuations among tartar samples in both tartar groups.
Ten out of 37 miRNAs showed significant different levels between two sample groups of which 9 miRNAs were decreased and one miRNA, miR-16-3p, increased in ancient tartar samples as compared to modern ones (Figure 2).
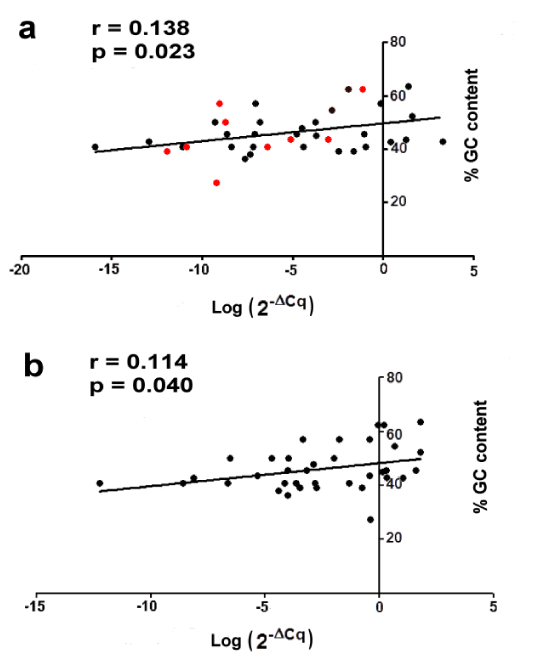
A positive correlation was observed between GC content and miRNA levels when ancient or modern samples were analyzed separately (Figure 3a,b). This suggests that miRNAs with higher GC content also show higher miRNA levels confirming the results described by Keller et al. in archaeological human specimens [10]. To test whether this behavior was the same for 9 decreased miRNAs in ancient tartar as compared to modern ones, we have compared these miRNAs between two tartar groups. The analysis showed that lower miRNA expression in ancient samples was not associated to lower GC content (red points in figure 3a).
Discussion
The major area of activity of forensic odontology is the identification of human beings and this is more so in case of natural or man-made disasters. In all these cases the most resistant human tissues are the teeth that survive to several traumatic forces [27]. Among various methods employed in forensic odontology include also the analysis of teeth DNA that it is generally highly fragmented and present only in minute amounts [28]. Other biomarkers, such as RNAs, have also been proposed and employed in forensic science [29-31] and, in addition, with advances in molecular biological techniques, novel approaches based on miRNA profiling have been developed for a possible forensic application [32]. For example, the circulating miRNAs have been considered as potential tool for identification of sex [33], body fluids [34,35] or organ tissue types [36].
Since recent investigations on archeological dental calculus have demonstrated that this structure contains well preserved saliva-derived biomolecules for extended periods of time [37], we have evaluated the long-term stability of miRNAs in tartar matrix comparing 40 saliva-derived miRNAs in archaeological and non-archaeological tartar samples.
After RT-PCR analysis, 3 out of 40 miRNAs were not detected in both ancient and modern samples, as well as in saliva samples of 10 out of 30 healthy volunteers chosen at random. An explanation of this phenomenon could be the lack of expression of these three miRNAs in both ancient and modern Puglia population since the choice of 40 miRNAs was based on literature data that have considered heterogeneous populations by race and geographical area. Few studies have considered the miRNA profiles in multiethnic populations. Huang et al. have found different expression of 33 miRNA in two HapMap lymphoblastoid cell lines representing non-African and African populations, respectively [38], and more recently Rawlings-Goss et al. have identified several miRNA genetic variants among African, Asian and European populations [39]. However additional investigations are needed on this topic.
Ten of remaining 37 miRNAs had different levels between two tartar sample groups. In particular 9 miRNAs were down-regulated while only one was up-regulated in ancient samples than modern ones. There may be several explanations for the observed discrepancies in our findings.
First, decreased miRNA levels in ancient tartar could be explained by degradation due to long-time of conservation in tartar matrix, however we found no correlation between GC content, used as biomarker of degradation, and miRNA levels. Second, it is known that miRNAs play key role in the loss of periodontal tissue integrity and an alternative hypothesis of the miRNA dysregulation could be the presence of the periodontal diseases in ancient individuals [40]. Recent studies have shown that miR-30b-3p, let-7i-5p, miR-125a-5p and miR-203c-3p were down-regulated, while miR-146a-3p, miR-20a-3p and miR-20a-5p were up-regulated in gum inflammation [41,42]. The same miRNAs have been analyzed in tartar samples by us, but no statistically significant differences between ancient and modern samples were found. Therefore, other plausible reasons could explain this phenomenon.
In the last years some studies have examined diet-miRNA interplay showing that some food components or dietary preferences may modulate serum miRNA profiles that in turn influence several biological processes [43]. For example, Ryu et al. have revealed that circulating miR-92a expression was positively influenced by abnormal dietary zinc levels in blood [44]. In agreement with these results, in a recent archeological study we found that high zinc levels in bone samples of a subject lived in the 14th century in Canosa (Puglia) were positively associated to high miR-92a levels in tartar samples [45]. Consequently, it is possible hypothesize that changes of the miRNA expression in ancient samples could be due to dietary shifts between ancient and modern populations. However, at present, limited information is available on the influence of specific food on the change of miRNA expression in human as well as it is still difficult today to have meaningful information about diet habits in ancient Puglia populations [46]. Therefore, it is necessary to focus on developing a gradual strategy for identifying specific miRNAs for different dietary compounds or eating habits.
Conclusion
Our study suggests that it is possible to recover well preserved miRNAs in the tartar matrix increasing the sustainability of dental calculus research. Although the cause of different levels of some miRNAs between ancient and modern tartar samples has not been explained because it is influenced by several factors, our findings show that tartar matrix is a barrier that protects the miRNAs from several sources of degradation such as water, enzymes and microorganism for extended periods of time. Therefore the analysis of the saliva-derived miRNAs in dental calculus could provide further valuable information to establish postmortem individual identification of unknown human remains, especially for unidentified subjects following mass calamities, fire or explosions of which the teeth are the only human tissue available for analysis. It must also be underlined that our study is a pilot study and in future further investigations are needed to establish if miRNA-based identification procedures in tartar samples are a reliable source.
We thank Dr. Chiara Marini and her colleagues (private Dental Clinic of Bari, Italy) for collecting modern dental calculus samples. This research did not receive any specific grant from funding agencies in the public, commercial, or non-for-profit sectors.
- Divakar KP (2017) Forensic odontology: the new dimension in dental analysis. Int J Biomed Sci 13: 1-5. Link: https://bit.ly/2Il6SHV
- Prajapati G, Sarode SC, Sarode GS, Shelke P, Awan KH, et al. (2018) Role of forensic odontology in the identification of victims of major mass disasters across the world: A systematic review. PLoS One 13: e0199791. Link: https://bit.ly/2tiVqCg
- Reesu GV, Augustine J, Urs AB (2015) Forensic considerations when dealing with incinerated human dental remains. J Forensic Leg Med 29: 13-17. Link: https://bit.ly/2Byh49V
- Sweet DJ, Sweet CH (1995) DNA analysis of dental pulp to link incinerated remains of homicide victim to crime scene. J Forensic Sci 40: 310-314. Link: https://bit.ly/2GCWf0w
- Higgins D, Austin JJ (2013) Teeth as a source of DNA for forensic identification of human remains: a review. Sci Justice 53: 433-441. Link: https://bit.ly/2RT5gnX
- Mann AE, Sabin S, Ziesemer K, Vagene AJ, Schroeder H et al. (2018) Differential preservation of endogenous human and microbial DNA in dental calculus and dentin. Sci Rep 8: 9822. Link: https://go.nature.com/2SxNUCA
- Poor VS, Lukacs D, Nagy T, Racz E, Sipos K (2016) The rate of RNA degradation in human dental pulp reveals post-mortem interval. Int J Legal Med 130: 615-619. Link: https://bit.ly/2MZnMu2
- Keller A, Kreis S, Leidinger P, Maixner F, Ludwig N, Backes C et al. (2017) miRNAs in ancient tissue specimens of the Tyrolean Iceman. Mol Biol Evol 34: 793-801. Link: https://bit.ly/2GDWZlS
- Rapado-Gonzalez O, Majem B, Muinelo-Romay L, Alvarez-Castro A, Santamaría A et al. (2018) Human salivary microRNAs in cancer. J Cancer 9: 638-649. Link: https://bit.ly/2SINCrr
- Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH et al. (2010) The microRNA spectrum in 12 body fluids. Clin Chem 56: 1733-1741. Link: https://bit.ly/2UVyVyV
- Yeri A, Courtright A, Reiman R, Carlson E, Beecroft T, et al. (2017) Total extracellular small RNA profiles from plasma, saliva, and urine of healthy subjects. Sci Rep 7: 44061. Link: https://bit.ly/2theUa3
- Park NJ, Zhou H, Elashoff D, Henson BS, Kastratovic DA et al. (2009) Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res 15: 5473-5477. Link: https://bit.ly/2tjPwRc
- Palazzo AF, Lee ES (2015) Non-coding RNA: what is functional and what is junk? Front Genet 6: 2. Link: https://bit.ly/2GDwDAu
- Mestdagh P, Hartmann N, Baeriswyl L, Andreasen D, Bernard N et al. (2014) Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods 11: 809-815. Link: https://bit.ly/2RRIqNH
- Beck LA (1995) Standards for data collection from human skeletal remains. Am J Human Biol 7: 672-672. Link: https://bit.ly/2GDIYVf
- Kirsanow K, Burger J (2012) Ancient human DNA. Ann Anat 194: 121-132. Link: https://bit.ly/2I7RCxS
- Rohland N, Hofreiter M (2007) Ancient DNA extraction from bones and teeth. Nat Protoc 2: 1756-1762. Link: https://bit.ly/2UOfUOS
- Yang DY, Watt K (2005) Contamination controls when preparing archaeological remains for ancient DNA analysis. J Archaeol Sci 32: 331-336. Link: https://bit.ly/2tkPwR4
- Cavallini A, Messa C, Pricci M, Caruso ML, Barone M et al. (2002) Distribution of estrogen receptor subtypes, expression of their variant forms, and clinico-pathological characteristics of human colorectal cancer. Dig Dis Sci 47: 2720–2728. Link: https://bit.ly/2I8H1T9
- Rotelli MT, Di Lena M, Cavallini A, Lippolis C, Bonfrate L et al. (2015) Fecal microRNA profile in patients with colorectal carcinoma before and after curative surgery. Int J Colorectal Dis 30: 891-898. Link: https://bit.ly/2RSdzAF
- Kakimoto Y, Tanaka M, Kamiguchi H, Ochiai E, Osawa M et al. (2016) MicroRNA stability in FFPE tissue samples: dependence on GC content. PLoS one 11: e0163125. Link: https://bit.ly/2tgbR1Y
- Mori N, Mizuno D, Goto S (1978) Increase in the ratio of 18S RNA to 28S RNA in the cytoplasm of mouse tissues during aging. Mech Ageing Dev 8: 285–297. Link: https://bit.ly/2BwT4Uu
- Cardin SE, Borchert GM (2017) Viral microRNAs, host microRNAs regulating viruses, and bacterial microRNA-like RNAs. Methods Mol Biol 1617: 39-56. Link: https://bit.ly/2I9qsXc
- Warinner C, Rodrigues JF, Vyas R, Trachsel C, Shved N et al. (2014) Pathogens and host immunity in the ancient human oral cavity. Nat Genet 46: 336-344. Link: https://bit.ly/2TOrtWk
- Kemp BM, Winters M, Monroe C, Barta JL (2014) How much DNA is lost? Measuring DNA loss of short-tandem-repeat length fragments targeted by the PowerPlex 16 system using the Qiagen MinElute Purification Kit. Hum Biol 86: 313-329. Link: https://bit.ly/2MZ7jpG
- Yoshizawa JM, Wong DT (2013) Salivary microRNAs and oral cancer detection. Methods Mol Biol 936: 313-324. Link: https://bit.ly/2SIjDjy
- Berketa J, Higgins D (2017) Stabilization of dental structures of severely incinerated victims at disaster scenes to facilitate human identification. J Forensic Leg Med 51: 45-49. Link: https://bit.ly/2Bu3gxe
- Rohland N, Glocke I, Aximu-Petri A, Meyer M (2018) Extraction of highly degraded DNA from ancient bones, teeth and sediments for high-throughput sequencing. Nat Protoc 13: 2447-2461. Link: https://go.nature.com/2I9pg6p
- Lindenbergh A, de Pagter M, Ramdayal G, Visser M, Zubakov D et al. (2012) A multiplex (m)RNA-profiling system for the forensic identification of body fluids and contact traces. Forensic Sci Int Genet 6: 565-577. Link: https://bit.ly/2GA5nmy
- Zubakov D, Kokshoorn M, Kloosterman A, Kayser M (2009) New markers for old stains: stable mRNA markers for blood and saliva identification from up to 16-year-old stains. Int J Legal Med 123: 71–74. Link: https://bit.ly/2N2Jg9i
- Tu C, Du T, Shao C, Liu Z, Li L, Shen (2018) Evaluating the potential of housekeeping genes, rRNAs, snRNAs, microRNAs and circRNAs as reference genes for the estimation of PMI. Forensic Sci Med Pathol 14: 194-201. Link: https://bit.ly/2E6x7h6
- Sirker M, Fimmers R, Schneider PM, Gomes I (2017) Evaluating the forensic application of 19 target microRNAs as biomarkers in body fluid and tissue identification. Forensic Sci Int Genet 27: 41-49. Link: https://bit.ly/2tiCYtt
- Cui C, Yang W, Shi J, Zhou Y, Yang J, Cui Q, Zhou Y (2018) Identification and analysis of human sex-biased microRNAs. Genomics Proteomics Bioinformatics 16: 200-211. Link: https://bit.ly/2I5Avg0
- Sauer E, Reinke AK, Courts C (2016) Differentiation of five body fluids from forensic samples by expression analysis of four microRNAs using quantitative PCR. Forensic Sci Int Genet 22: 89-99. Link: https://bit.ly/2DGx4Hp
- O Leary KR, Glynn CL (2018) Investigating the isolation and amplification of microRNAs for forensic body fluid identification. Microrna 7: 187-194. Link: https://bit.ly/2TQn6dn
- Sauer E, Extra A, Cachée P, Courts C (2017) Identification of organ tissue types and skin from forensic samples by microRNA expression analysis. Forensic Sci Int Genet 28: 99-110. Link: https://bit.ly/2WPJXaC
- Velsko IM, Overmyer KA, Speller C, Klaus L, Collins MJ et al. (2017) The dental calculus metabolome in modern and historic samples. Metabolomics 13: 134. Link: https://bit.ly/2I36UUq
- Huang RS, Gamazon ER, Ziliak D (2011) Population differences in microRNA expression and biological implications. RNA Biol 8: 692-701. Link: https://bit.ly/2Bx1Guy
- Rawlings-Goss RA, Campbell MC, Tishkoff SA (2014) Global population-specific variation in miRNA associated with cancer risk and clinical biomarkers. BMC Med Genomics 7: 53. Link: https://bit.ly/2GmTEIH
- Irwandi RA, Vacharaksa A (2016) The role of microRNA in periodontal tissue: A review of the literature. Arch Oral Biol 72: 66-74. Link: https://bit.ly/2SMOlrK
- Luan X, Zhou X, Trombetta-eSilva J, Francis M, Gaharwar AK et al. (2017) MicroRNAs and periodontal homeostasis. J Dent Res 96: 491-500. Link: https://bit.ly/2SvckfV
- Riaz Rajoka MS, Jin M, Haobin Z, Li Q, Shao D, Huang Q et al. (2018) Impact of dietary compounds on cancer- related gut microbiota and microRNA. Appl Microbiol Biotechnol 102: 4291-4303. Link: https://bit.ly/2DGxj5h
- Cui J, Zhou B, Ross SA, Zempleni J (2017) Nutrition, microRNAs, and human health. Adv Nutr 8: 105-112. Link: https://bit.ly/2BwR6DT
- Ryu MS, Langkamp-Henken B, Chang SM, Shankar MN, Cousins RJ (2011) Genomic analysis, cytokine expression, and microRNA profiling reveal biomarkers of human dietary zinc depletion and homeostasis. Proc Natl Acad Sci USA 108: 20970–20975. Link: https://bit.ly/2BwR6DT
- Cavallini A (2018) Che patologie aveva: le indagini chimiche e biomolecolari. In: Una finestra sulla storia. Un cavaliere a Castiglione tra angioini e aragonesi (eds. Perrino G and Sublimi Saponetti S) - Quaderni della Sezione Sudest Barese Press, Conversano (BA), Italy. 95-98.
- Renna M, Rinaldi VA, Gonnella M (2015) The Mediterranean diet between traditional foods and human health: the culinary example of Puglia (Southern Italy). Int J Gastron Food Sci 2: 63-71. Link: https://bit.ly/2N1Afxt
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
