Annals of Systems Biology
SARS-Cov-2 Systems Biology
José Díaz*
Cite this as
Díaz J (2020) SARS-Cov-2 Systems Biology. Ann Syst Biol 3(1): 029-032. DOI: 10.17352/asb.000009The aim of this mini review is to analysis the advances in the research of the SARS-CoV-2 molecular structure and pathogenesis from a systems biology approach.
Introduction: Experimental analysis of the interaction of viral and host proteins, or interactome, by Gordon and collaborators has been a fundamental contribution to understand the form in which SARS-CoV-2 virus takes control of the host molecular network to produce new virions and propagate the infection. This result allows the construction of the viral network representation of the interactome and its statistical analysis. Formulation of network models of this interactome is a basic tool to identify drug targets capable of interrupt the viral replication cycle, and for the design of novel therapeutic agents.
Discussion: SARS-CoV-2 is a free-scale hierarchical modular structure in which the open reading frame 8 protein, nucleocapside protein (N), and nonstructural protein 7 (Nsp7) are the central hubs. This kind of organization confers an extra level of complexity to this molecular network allowing it to resist the attack of drugs on single nodes. However, simultaneous suppression of these three hubs can effectively disrupt the network.
Conclusions: Systems biology approach to the analysis of the SARS-CoV-2 interactome reveals the existence of six nodes that belong to high modularity classes (open reading frame 8 protein (orf8), Membrane protein (M), open reading frame 9b protein (orf9b), Nucleocapside protein (N), open reading frame 10 protein (orf10), Envelope protein (E), open reading frame 6 protein (orf6), open reading frame 7 protein (orf7) and Spike protein (S)) and control the flow of information during infection. These proteins are possible targets for new drugs, principally for antagonist of the three proteins orf8, M and Nsp7 that are the main bottlenecks throw which most of the information required for the production of new virus must flow. A therapeutic attack on these hubs can increase the probability to defeat viral infection as an alternative to a vaccine.
Abbreviations
SARS-CoV-2: Severe Acute Respiratory Syndrome Coronavirus 2; COVID19: Coronavirus Disease 2019; ACE2: Angiotensin-Converting Enzyme 2; Ang II: Angiotensin II; Ang (1-7): Angiotensin (1-7); Nsp: Nonstructural protein: RTC: Replication-Transcription Complex; AI: Artificial Intelligence; ODE: Ordinary Differential Equation; orf: Open Reading Frame; Nsp: Nonstructural protein; E: Envelop protein; S: Spike protein; M: Membrane protein; N: Nucleocapside protein
Introduction
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) virus is an intracellular parasite whose replication cycle depends on host cell structures and functions; in particular, it uses the translational apparatus of different types of infected cells to express its proteins [1]. SARS-CoV-2 causes the COVID19 (Coronavirus Disease 2019), which has infected around 21,000,000 persons worldwide since the end of 2019 and killed about 776,000 of them. There are not therapeutic drugs neither an effective vaccine to defeat SARS-CoV-2 infection [2]. .
SARS-CoV-2 virion is formed by four proteins: Spike (S), Envelope (E), Membrane (M) and Nucleocapside (N) that enclose the virus genome [3]. In normal cells, the surface receptor Angiotensin-Converting Enzyme 2 (ACE2), highly abundant in the lung alveolar type II cells, converts the molecules of Angiotensin II (Ang II) into Angiotensin (1-7) (Ang (1-7)) [4]. However, when SARS-CoV-2 virion infects the organism, ACE2 binds with high affinity to the S protein, and forms a molecular complex that begins the process of fusion of the virion envelope and the host cell membrane. The nucleocapside containing the viral genome in then released into the host cytoplasm [5].
SARS-CoV-2 genome consists of a positive-sense nonsegmented single stranded mRNA ((+)ssRNA) of 30 kb. The open reading frames 1a (orf1a) and 1b (orf1b) are located near the untranslated region 5’ (5’UTR) of the positive single stranded RNA ((+)ssRNA) and they code for the polyproteins pp1a and pp1ab. The process of translation occurs in the cytoplasm and produces a set of viral polyproteins whose maturation results in 11 nonstructural proteins (Nsp) from the orf1a segment (Nsp1 to Nsp11) and 5 nonstructural proteins from the orf1b segment (Nsp12 to Nsp16). Nsp proteins form the replication-transcription complex (RTC) in a double-membrane vesicle where a set of nested subgenomic minus-strands of RNA ((-)sgRNA) are synthesized in a process of discontinuous transcription. These (-) sgRNAs serve as the templates for the production of subgenomic mRNAs from which the structural proteins E, M, N and S, together with the accessory proteins orf 3a, orf6, orf7a, orf7b, orf8, orf9b, orf9c and orf10 are synthesized [2,6,7]. SARS-CoV-2 uses the host translational machinery to redirect it to viral protein synthesis and replication, while cellular mRNA translation is inhibited [1]. Viral proteins are inserted into the host molecular machinery to modify and redirect a great number of host cell functions towards the production of more virus particles [8,9]. Experimental analysis of the interaction of viral and host proteins, or interactome, by Gordon and collaborators [10] has been a fundamental contribution to understand the form in which SARS-CoV-2 virus takes control of the host molecular network to produce new virions and propagate the infection. Gordon and collaborators [10] cloned, tagged and expressed 26 viral proteins in human cells using affinity- purification mass spectrometry to identify the human proteins physically associated with each other. They found around 332 SARS-CoV-2-human protein-protein interactions that form the virus interactome. This result allows the construction of the viral network representation of the interactome and its statistical analysis. Formulation of network models of this interactome is a basic tool to identify drug targets capable of interrupt the viral replication cycle, and for the design of novel therapeutic agents [2,11-13].
Discussion
Systems Biology uses network theory as a tool to understand the organization and dynamics of complex systems [14]. A theoretical approach to biological networks structure and function allows the integration of disperse experimental data in a coherent model of the spatio-temporal dynamics of interconnected cellular processes [2]. The number of nodes, the number of connections of each node to his neighbours, and the distribution of these connections in the network determine its complexity, structure and dynamical properties [15].
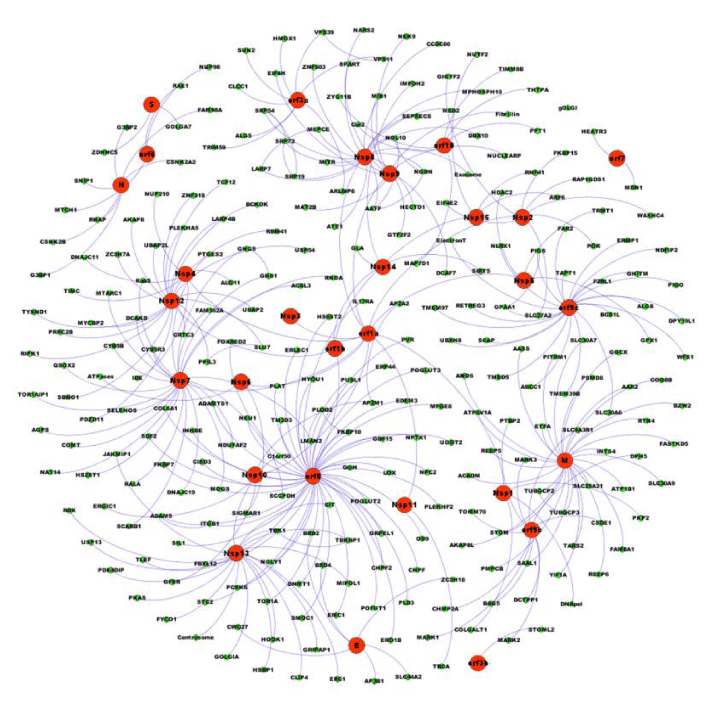
In the particular case of SARS-CoV-2 virus, the analysis of the statistical properties of the undirected network model (Figure 1) of its interactome [2,10,15] shows that the degree distribution of its 332 proteins or nodes is a decaying exponential with great number of nodes with few connections and a low number of nodes with degree above 20. A model of the Calu-3-specific human-SARS-CoV-2 interactome, with 4,123 nodes, also shows a degree decaying exponential distribution [15]. Both results suggest that the SARS-CoV-2 network has a scale-free structure. In particular, orf8, M, Nsp7, orf9c, Nsp12 and Nsp13 proteins are the most connected nodes in the network and can be considered as hubs [2]. Three hubs from this set control approximately 102 viral-host processes [2] as shown in Figure 1: orf8 (47 links), M (29 links) and Nsp7 (26 links). Hubs identification is of great importance because these highly connected nodes control the flow of information in the infected cell, indicating that orf8, M and Nsp7 are the main organizers of the SARS-CoV-2 virus activity during infection and the possible source of the virus pathogenicity [2,13,15].
All networks have a structure determined by nodes connectivity and links distribution. In random nets connectivity and links distribution are arbitrary and the degree distribution follows a binomial distribution. However, every binomial distribution can be approximated with a Poisson distribution that is also a decaying exponential one. In consequence, a decaying exponential degree distribution is not a sufficient condition to assure that SARS-CoV-2 is not a random network, i.e., that in every case of infection viral proteins are inserted at random in the host cell without a specific coordinated function.
Centrality measures are necessary to determine if an undirected network is random or not. Different measures have been employed to investigate the structure of the SARS-CoV-2 interactome: clustering coefficient, closness centrality, betweeness centrality and modularity class, load centrality, information centrality and Page Rank index [2,15]. From these set of centrality measures, modularity is the key to determine if the host cell-virus interactome has any kind of nonrandom structure.
Modularity is the fraction of links that fall within a cluster minus the expected fraction if links were distributed at random, and indicates the nodes that are more densely connected between them than with the rest. In a nonrandom network, modularity has a value between 0 and 1 and reveals clues about the structure and the vulnerable spots of the network. A common method used for community detection is the Louvain method [2,16], from which the modularity of the undirected SARS-CoV-2 was calculated as approximately 0.85 that is far from the negative value for a random network. Furthermore, in the Gordon et al. interactome [10], 21 clusters were detected for which the number of nodes distributed between them is larger than the expected number due to random. The viral proteins orf8, M, orf9b, N, orf10, E, orf6, orf7 and S also belong to high modularity classes. These results suggest that SARS-CoV-2 is not a random network but a free-scale modular hierarchical structure in which orf8, N, and Nsp7 are the central hubs [2]. This kind of organization confers an extra level of complexity to the SARS-CoV-2 molecular network allowing it to resist the attack of drugs on single nodes. However, simultaneous suppression of these three hubs could effectively disrupt the network organization and stop the viral replication cycle [17]. These results reveal the complexity of the SARS-CoV-2 molecular network, and open the possibility to understand the factors that determine the pathogenicity of the virus [2,13], and how can they be modified to decrease it [2,13,15,18].
In silico analysis of the structure and dynamics of the SARS-CoV-2 network can be of aid in the identification of drug targets and in the design of novel medicaments against COVID19. Artificial Intelligence (AI) and bioinformatics methods have been used for this purpose with great success [19,20].
CoV-responsive human genes and their functional roles were indentified based on available genomic data and on the basis of both the relative synonymous codon usage (RSCU)-based correlation of viral genes with human genes and differential gene expression analysis. From this analysis, potential drugs for COVID-19 treatment based on these genes were predicted [21]. Furthermore, experimental data from Gordon, et al. [10], suggest that drug attack on orf8, N, and Nsp7 can block the effect of these three hubs on downstream nodes [2]. However, there are not therapeutic drugs that can hit directly these proteins, although there are FDA approved drugs than block their downstream activity. For example, orf8 is a target of Rapamycin, which also targets Nsp2 and N. Rapamycin blocks Tor1a activity in the quality control of protein folding in the Endoplasmic Reticulum (ER), disrupting the effects of orf8 [22,23]. Unfortunately, Rapamycin has strong immunosuppressant effects which turn it inadequate for the treatment of the COVID19 disease [21]. It is necessary more research to design drugs that target directly orf8, M and Nsp7 as a complement or an alternative to a vaccine.
Conclusion
Systems biology approach to the analysis of the SARS-CoV-2 interactome reveals that is a modular free-scale hierarchical network in which six nodes orf8, M, orf9b, N, orf10, E, orf6, orf7 and S that belong to high modularity classes control the flow of information during infection. These proteins are possible targets for new drugs, principally for antagonist of the three proteins orf8, N, and Nsp7 that are the main bottlenecks throw which most of the information required for the production of new virus must flow. A therapeutic attack on these hubs can increase the probability to defeat viral infection as an alternative or a complement to a vaccine.
The next step is the formulation of an Ordinary Differential Equations (ODEs) model of the SARS-CoV-2 interactome to know in deep the dynamical behavior of this complex network. This kind of modeling is necessary because viral protein insertion is a perturbation that distorts the original host cell phase space topology. Probably, viral proteins produce some type of bifurcation, with a restructuration of the original fixed points, which leads the overall host cell dynamics to the production of new virus [24]. Characterization of the kind of bifurcation involved in the infection by SARS-CoV-2 is a necessary step to know the factors that determine the pathogenicity of the virus [2], and how they can be modified with specific drugs to decrease it.
However, there is a lack of the quantitative information necessary to propose an ODEs-based continuous model for the analysis of the spatio-temporal dynamics of the infection due to the novelty of this virus species.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
I thank CIDC colleagues for their constructive comments and suggestions. I also thank Erika Juarez Luna for logistical support.
- Nakagawa K, Lokugamage KG, Makino S (2016) Viral and Cellular mRNA Translation in Coronavirus-Infected Cells. Advances in Virus Research 96: 2016. Link: https://bit.ly/3haJKrC
- Diaz J (2020) SARS-CoV-2 Molecular Network Structure. Front Physiol 10. Link: https://bit.ly/3k18QuM
- McBride R, Fielding BC (2012) The Role of Severe Acute Respiratory Syndrome (SARS)-Coronavirus Accessory Proteins in Virus Pathogenesis. Viruses 4: 2902-2923. Link: https://bit.ly/2RcN0YW
- Magalhaes GS, Rodrigues-Machad MG, Motta-Santos D, Campagnole-Santos MJ, Santos RAS (2020) Activation of Ang-(1-7)/Mas Receptor Is a Possible Strategy to Treat Coronavirus (SARS-CoV-2) Infection. Front Physiol 11: 730. Link: https://bit.ly/2ZqtFI9
- Letko M, Marzi A, Munster V (2020) Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nature Microbiology 5: 562–569. Link: https://bit.ly/3bKmL5J
- Sevajol M, Subbisi L, Decroly E, Canard B, Imbert I (2014) Insights into RNA synthesis, capping, and proofreading mechanisms of SARS-coronavirus. Virus Research. Link: https://bit.ly/3m7Gr86
- Dongwan K, Joo-Yeon L, Jeong-Sun Y, Won KJ, Narry KV, et al. (2020) The architecture of SARS-CoV-2 transcriptome. BioRxiv preprint. Link: https://bit.ly/3hgI4wS
- Masters PS (2006) The Molecular Biology of Coronaviruses. Advances in Virus Research 66: 193-292. Link: https://bit.ly/3jXCReW
- Wu A, Peng Y, Huang B (2020) Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host & Microbe. 27: 325-338. Link: https://bit.ly/2ZpGJh1
- Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, et al. (2020) A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing. BioRxiv preprint. Link: https://bit.ly/3m8I9GA
- Scavone C, Brusco S, Bertini M, Sportiello L, Rafaniello C, et al. (2020) Current pharmacological treatments for COVID-19: what's next? British Journal of Pharmacology. Link: https://bit.ly/2RpQMyp
- Sadegh S, Matschinske J, Blumenthal DB, Galindez G (2020) Exploring the SARS-CoV-2 virus-host-drug interactome for drug repurposing. Nature Communications 11: 3518. Link: https://bit.ly/3m8e21T
- Messina M, Giombini E, Agrati C, Francesco Vairo F (2020) COVID‑19: viral–host interactome analyzed by network based‑approach model to study pathogenesis of SARS‑CoV‑2 infection. J Transl Med 18:233. Link: https://bit.ly/2ZG0W2r
- Breitling R (2010) What is systems biology? Front Physiol 1: 9. Link: https://bit.ly/3maH2WT
- Kumar N, Mishra B, Mehmood A, Athar M, Mukhtar MS (2020) Integrative Network Biology Framework Elucidates Molecular Mechanisms of SARS-CoV-2 Pathogenesis. BioRxiv preprint. Link: https://bit.ly/2ReG8Ku
- Brandes U, Delling D, Gaertler M, Gorke R, Hoefer M, et al. (2008) On Modularity Clustering. IEEE Transactions on Knowledge and Data Engineering. IEEE 20: 172–188. Link: https://bit.ly/3heex6N
- Cohen R, Erez K, ben-Avraham D, Havlin S (2000) Resilience of the Internet to random breakdowns. Phys Rev Lett 85: 4626. Link: https://bit.ly/3k4U0TY
- Zhou Y, Hou Y, Shen J, Huang Y, Martin W, et al. (2020) Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discovery 6:14. Link: https://bit.ly/3ii8788
- Lalmuanawma S, Hussain J, Chhakchhuak L (2020) Applications of machine learning and artificial intelligence for Covid-19 (SARS-CoV-2) pandemic: A review. Chaos Solitons Fractals 139: 110059. Link: https://bit.ly/2RbPzdL
- Robson B (2020) Computers and viral diseases. Preliminary bioinformatics studies on the design of a synthetic vaccine and a preventative peptidomimetic antagonist against the SARS-CoV-2 (2019-nCoV, COVID-19) coronavirus. Computers in Biology and Medicine 119: 103670. Link: https://bit.ly/35mE7El
- Nambou K, Anakpa M (2020) Deciphering the co-adaptation of codon usage between respiratory coronaviruses and their human host uncovers candidate therapeutics for COVID-1. Infection, Genetics and Evolution 85: 104471. Link: https://bit.ly/2Fs08Xl
- Omarjeea L, Janinc A, Perrot F, Laviollee B, Meilhac O, et al. (2020) Targeting T-cell senescence and cytokine storm with rapamycin to prevent severe progression in COVID-19. Clin Immunol 216: 108464.Link: https://bit.ly/33aMYq2
- Mannick JB, Morris M, Hockey HP, Guglielmo Roma G, et al. (2018) TORC1 inhibition enhances immune function and reduces infections in the elderly. Sci Transl Med 10: eaaq1564. Link: https://bit.ly/3hpjvxT
- Bensussen A, Torres-Sosa C, Gonzalez RA, Díaz J (2018) Dynamics of the Gene Regulatory Network of HIV-1 and the Role of Viral Non-coding RNAs on Latency Reversion Front Physiol 9:1364. Link: https://bit.ly/2FjyEU6
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
