Annals of Systems Biology
T-cell malignancies pathogenesis, a system hematology/oncology
Jalal Naghinezhad1*, Jalil Rohani2, Shahriyar Gholizadeh2, Sajjad Ehtiati3, Jamshid Gholizadeh4 and Hossein Ayatollahi1
2Department of Biotechnology, Faculty of Medicine, Mashhad University of Medical Sciences, Mashhad, Iran
3Department of Clinical Biochemistry, Faculty of Medicine, Mashhad University of Medical Sciences, Mashhad, Iran
4Department of Clinical Immunology, Inflammation and inflammatory research center, Faculty of Medicine, Mashhad University of Medical Sciences, Mashhad, Iran
Cite this as
Naghinezhad J, Rohani J, Gholizadeh S, Ehtiati S, Gholizadeh J, et al. (2019) T-cell malignancies pathogenesis, a system hematology/oncology. Ann Syst Biol 2(1): 001-007. DOI: 10.17352/asb.000002Human T-cell leukemia/lymphoma is one of the most aggressive hematologic malignancy, which associated with poor prognosis. T-cell prolymphocytic leukemia (TPLL), Sezary syndrome, and acute T cell lymphocytic leukemia (T-ALL) are included to T-cell neoplasm. It is important to understand the alteration among expression of the genes toward malignant T cells. For acquiring more insights in oncogenesis molecular events, the systems hematology analysis was done. At first, the differentially expressed genes (DEGs) were taken pairwise among the four sample sets of T-PLL .vs normal T-cell, Sezary syndrome .vs normal T-cell, and ATLL .vs normal T-cell. Then, the protein-protein interaction networks (PPINs) were reconstructed via using the hub genes. Finally, the pathways of cells proliferation and transformation were identified TPLL and Sezary syndrome and ATLL. Among all differentially expressed gene (DEG), 1538 gene has log fold change above 2 or under -2. The hub genes among DEGs were found. The results demonstrated, oncogenesis genes associated with proliferation, evade apoptosis and inflammation were significantly overexpressed in T-cell disorders. comparative investigation gene expression between T-PLL .vs normal T-cell, Sezary syndrome .vs normal T-cell, and ATLL .vs normal T-cell showed despite inflammatory genes such AP-1, AIF, NF-kB were overexpressed in T-cell disorders but in ATL it occurred significantly and important pathways like PI3K/Akt/mTOR are activated in T-cell disorders especially in ATL.
Introduction
Recent advances in cellular and molecular biology producing an excessive volume of complicated datain an interaction manner between hormones, neurotransmitters, eicosanoids, cytokines and genes. In addition, intra and extra protein-protein interaction network occurs among the cells in response to environmental triggers, which brings more intricacy Specify the outcome of an activity in a special tissue or organ to the surrounding microenvironment make it more intricacy. There are two main problems in reductionist molecular approach, the first one is producing information pollution and secondly, the systemic or physiological or physio-pathological interpretation are very hard. Therefore, using systemic approach in context of holistic program may help to understand the pathway which may produce particular consequences. Therefore, system biology programs might be helpful in such scientific situation.
T-cell lymphoproliferative disorders are a broad type of malignancy, which included several progressive cancers including T-PLL (T-cell prolymphocytic leukemia), sezary syndrome, and T-ALL.
T-PLL is one of the mature T-cell neoplasm, which accounts for 2% of all mature lymphocytic leukemia [1,2]. In this type of disease, there is post-thymic monoclonal prolymphocyte, which strongly expresses CD7, CD5, and CD2, and the lack of expression of TdT and CD1a [3,4]. Malignant cells have the normal size, high ratio of nuclear-to-cytoplasm, with a round to oval shape, and a single prominent nucleolus; the cytoplasm is agranular, with blebs [1]. Sezary syndrome is another T-cell neoplasm, which characterized by the triad phenomena. 1. Wide reddening of the skin (erythroderma), 2. Sezary cells (atypical T-lymphocytes) in peripheral blood, 3. Skin rushes. Furthermore, the most pertinent clinical aspect is erythroderma [5,6]. Generally, Sezary cells are mature monoclonal T-cell that expresses CD3+, CD4+ and CD8–; it may aberrantly not express T-cell antigens like CD7 [7].
Adult T cell leukemia/Lymphoma (ATLL) is viral (HTLV) associated T-cell neoplasm, which characterized by abnormal T-cell in peripheral blood [8]. Despite the HTLV-1 is endemic in some region, however, only the minority of infected subjects express ATLL (2-5%) and the majority of them remain asymptomatic [9]. ATL has a poor prognosis with a survival rate of very short around 11 months in lymphomatous and acute forms.
HTLV-1 is the first discovered oncovirus which bearing two oncogenic proteins, Tax and HBZ. Tax is a viral tans-activator, the immune-dominant protein of HTLV-1 which induces cell cycle and growth factors such as IL-2, IL-2Rα, GM-CSF. Therefore, HTLV-1 tax is necessary for cell transforming, while due to immunogenicity the virus decrement its expression by producing HBZ to escape from host immune system. Therefore, Tax inducing malignancy, but HBZ necessary for maintenance of the cancerous cells.
It is important to investigate and find hub genes that expressed in T-cell leukemia /lymphoma and analyze their role in this disease. So more investigation to find the hub DEGs expression among T-cell leukemia/lymphoma is necessary.
Material and Method
Gene expression microarray dataset
The gene expression profile of CD3 positive T-cells comprise a platform, GPL10558 Illumina HumanHT-12 V4.0 expression beadchip was obtained from NCBI Gene Expression Omnibus (GEO) database (Accession number: GSE107397 for T-PLL and sezary syndrome, GSE55851 for ATL with platform GPL10332 Agilent-026652 Whole Human Genome Microarray 4x44K v2).
Analysis of differentially expressed genes (DEGs)
The dataset was normalized by GEO2R. Differentially expression genes (DEGs) were identified through GEO detabase. In the same part fold change (FC) are also calculated. The DEGs were recognized between different data sets as three categories includes: 1) TPLL vs. normal T-cell, 2) sezary vs. normal T-cell, and 3) ATL vs. normal T-cell. Based on adjusted p-values <0.05 the DEGs was selected. The upregulated (positive logFC) and downregulated (negative logFC) genes recognized by Log FC. The heatmap was drawn by using package pheatmap in R 3.2.5.
Construction of PPIN and centrality analysis
For create the PPINs, we used the online STRING database. The cut off to analyze the PPINs were considered upper than 0.4 and we considered All interaction origins comprise physical and functional that may derive from co-expression, genomic context, high-throughput experiments, and previous. Then we used the Network Analyzer app in Cytoscape (3.7.0) for analyzation the PPINs. We calculate degree, closeness, and betweenness parameter by this software. The degree is a number of edges which are connected to each node. The closeness centrality is the shortest path between every two nodes. Another centrality parameter is the betweenness that is the number of their visiting during the crossing from all shortest paths. In the next step, each of the functional modules of T-PLL vs normal T-cell, Sezary syndrome vs normal T-cell, and ATL vs normal T-cell.
Finally, the Enrichr were utilized to enrich the identified hub DEGs in KEGG pathway to comprehend further intuition about communications. The hub genes of T-PLL vs normal T-cell, Sezary syndrome vs normal T-cell, and ATL vs normal T-cell were enriched in PI3K-Akt signaling, Ras signaling cancer, cancer, Pancreatic cancer, B cell receptor signaling cancer, and Adherence junction so the results confirm the cells proliferation, evade apoptosis, migration, and transformation due to change in aberrant signaling pathways.
Results
Correlation analysis of sample-sample
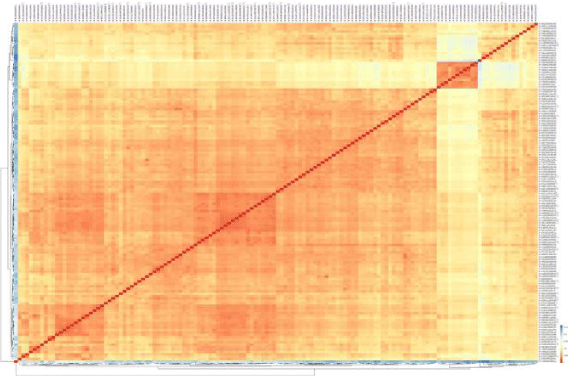
Correlation map (Figure 1) is the first figure of this paper and showed the distances of sample versus sample. Based on the sample correlation, this map compares the expression data and computed the results (expression of genes). The range of map color is from blue to red. Red shows no difference or higher correlation and blue indicates a low level of correlation (Figure 2).
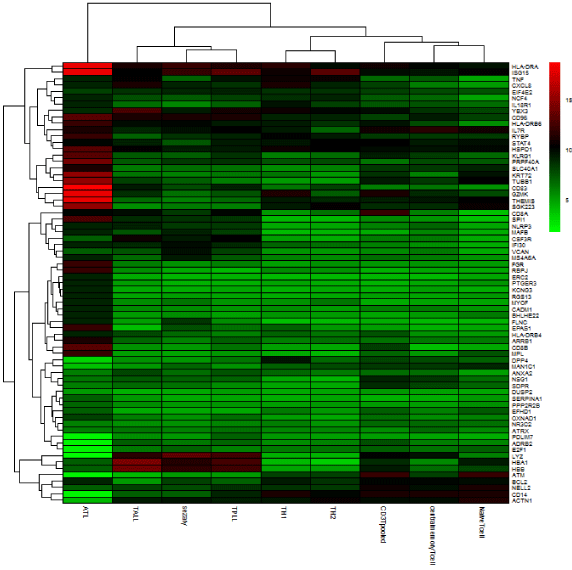
In the heat map, the first 70 genes (ordering by variance) belonging to each group were plotted. In this map, mean gene expression in each sample was calculated. Based on variance difference the top 70 genes among five sample sets were elected. Upregulated genes are specified as red and downregulated genes are identified as green.
Analysis of DEGs
Based on adjusted P-value <0.05 as the criteria, 4487, 1799, and 31580 genes with remarkable differential expressions were found for five categories of Normal vs. ATL, Normal vs. sezary syndrome, and Normal vs. T-PLL respectively.
The identification of hub genes and common genes
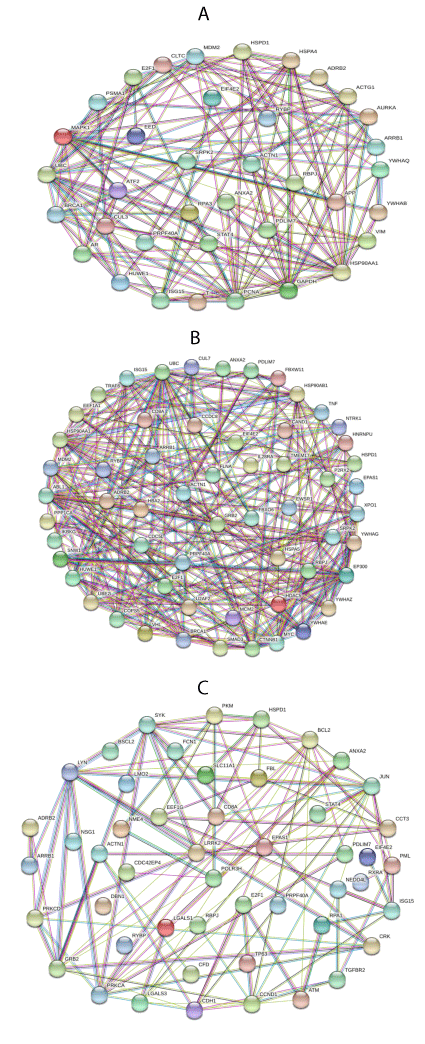
To examine the stock relationship between the determined hub genes and the development of ATL, sezary syndrome, and TPLL the PPINs were utilized. The node size demonstrates the degree. The node color was ordered according to the closeness centrality. The edge color showed betweenness. Red color demonstrates low closeness and blue shows high closeness centrality.
PPIN construction
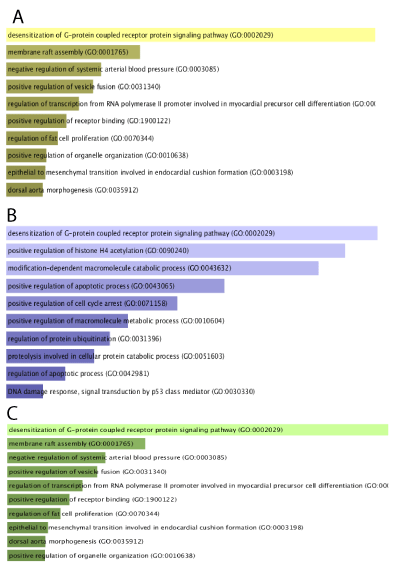
In gene ontology biological process (GO Biological Process) of enrichr website, each of the functional modules of TPLL vs. normal T-cell, sezary syndrome vs. normal T-cell, and ATL vs. normal T-cell were individually enriched. Afterward, based on inclusion of the gene, the meaningful modules were elected. Most of the nodes associated with cell proliferation, FC gamma receptors signaling and evade of apoptosis and inflammation. In addition, nucleotide-excision repair was upregulated (Figure 3).
The identification of functional modules and their ontology enrichment
Each of the functional modules of Normal vs. ATL, TPLL, and sezary syndrome were individually in gene ontology biological process (GO Biological Process) enriched. Most of the nodes associated with apoptosis and nucleotide-excision repair were upregulated, while the majority of the nodes associated with the negative regulation of cell differentiation were downregulated in ATL, TPLL, and sezary syndrome.
Discussion
The pathogenesis mechanism result from T-cell malignancies are not well recognized so far. Hub genes that involved in the development of the TPLL, ATL, and Sezary syndrome highlighted by network analysis. Sixty-two, fifty-eight, and forty-nine (Table 1,2) DEGs were found from TPLL Sezary syndrome, and ATL vs normal T-cells, respectively. These genes may involve in the pathogenesis pathway. Information extracted from KEGG demonstrated that these DEGs have a role in proliferation, evade of apoptosis, immune signaling pathways. DEGs between TPLL vs normal T-cell, Sezary vs normal T-cell, ATL vs. normal T-cell were enriched in GO Biologocal Process (Figure 4) and revealed the functions of these genes in positive regulation of transcription, desensitization G protein receptor signaling, and regulate apoptosis signaling. In addition, some genes upregulated that have a role in lymphocyte activation and induce inflammation such NF-kB and AP-1.
Some events exist that proposed to induce tumorigenesis, which comprised of inflammation, cell survival, apoptosis inhibition, DNA repair blocking through deregulation of the related gene expression. Lymphocyte activation is the essential aim of up- and down-regulated genes in ATLL progression. All of the three ways of lymphocyte activation are smart in ATL; I) NF-kappa B activation that lead to the upregulation of BCL2A1, II) Upregulation of MAP2K4 that result in activation of AP1 and III) Actuation of NFAT gene controlled from upregulation of CALM 1. Furthermore, to lymphocyte activation, NF-kappa B can stimulate cell survival, inflammatory responses, and apoptosis inhibition. Also, NF-kappa B is aroused by an increase of TLR5 and can be under direct influence of Tax protein [10].
One of the bold difference between ATL and other T-cell neoplasm is inflammation induced by HTLV-1. Some genes were overexpressed only in ATL that may stimulate through HTLV-1. HSPG2 takes part in the fundamental interaction between the HTLV-1 and the cell. On the other hand, HSPG2 works as a HTLV-1 receptor. The essential region of HSPG is endorepellin (C-terminal Domain V), which appended to the VEGFR2 upon endothelial cells and special integrin receptor (α2β1) [12]. CXCR3 was overexpressed significantly in just ATL, although in other malignant T-cell is slightly upregulated. This lead to enhances shift T-helper to TH-1 and induce inflammation and result in more progression of ATL [11]. Another important chemokine receptor that overexpressed in ATL is CXCR7, which regulates the interaction between endothelial cells and tumor cells in intra- and extravasation and exerts an anti-apoptotic action. Inflammatory proteins like allograft inflammatory factor 1 (AIF) is found in an immune cell in response to inflammatory cytokines like IL1 and INF gamma. In ATL upregulated significantly and lead to cell proliferation in response of inflammation. ISG 15 could be induced by HTLV-1 directly and upregulated common between T-PLL, ATL, sezary syndrome. ISG15 stimulate inflammation by activation of RIG1 and NF-kB.
In T-PLL and sezary syndrome development, the similar pathways to ATLL such mTOR and NF-kappa B pathways are influenced by different genes and lead to the same results. In T-PLL, TCL1A demonstrated the highest dysregulation. TCL1A as a booster of T-cell signaling input, its upregulation was associated with deregulations of TCR pathway modulators, suggesting a net improvement of antigen receptor and cytokine signaling in T-PLL. This included decreased expressions of the membranenegative-costimulatory CTLA4, reduce the suppressive T–T homotypic receptor SLAMF6, or of the T cell pro-apoptotic GIMAPs, andsignificant overexpression of TNF, also known to shape TCR signals. In addition, Upregulation of immunosuppressive CD83 result in more immune evasive properties [13]. In addition, TCL1A induce activation of NF-kB and result in activation and proliferation of malignant lymphocyte. Importantly this protein activates PI3K/Akt/mTOR pathway and interact with ATM. So has central role in T-PLL progression.
Over-activated pathways in TPLL, ATL, and sezary syndrome
PI3K/Akt/mTOR signaling pathway is activated in these malignancies in different ways. For example, in sezary syndrome, PTEN gene deletion leads to hyperactivation of this pathway and in ATL viral onco-proteins stimulate this pathway. This pathway is pivotal signaling pathway, which has a role in proliferation, survival, migration and evades of apoptosis [14]. This pathway regulates key proteins like MDM2, P27, P21, FKHR, caspase9, and NF-kB [15]. Activation of NF-kB lead to cell activation so has central role tumorgenesis. Furthermore, PI3K/AKT/mTOR regulate cell nutrition level and increase GLUT4 in the cell surface [16]. In term of anti-apoptotic function, this pathway downregulates BAD, BAX and upregulates BCL-2, BCL-xL, and MCL-1 [17]. In term of inflammation this pathway upregulates NF-kB. This pathway enhances cell proliferation through induce cap dependent translation by activation of mTOR.
Another important signaling pathway that activated in this type of malignancy is RAS signaling which has a pivotal role in proliferation and survival [18]. Ras signaling pathway activate important transcription factors like JUN, FOS, E2F.
Wnt/B catenin signaling pathway is critical pathway in pathogenesis of T-cell malignancies. This pathway has important role in tumorgenesis. This pathway activated from different rout. AKT can inhibit GSK3B (B catenin inhibitor) and activate this pathway. Besides Wnt binding to its receptor leads to activation of this pathway.
The JAK-STAT signaling pathway is an important signaling pathway that has a key role in T-cell malignancies, especially in T-PLL. This signaling pathway is associated with an important process like immunity, cell division, cell death, and tumor formation. This pathway can stimulate through different way such cytokine receptor and integrin.
Hub DEGs that contribute in TPLL, ATL, and sezary syndrome pathogenesis
A. downregulated hub genes: TP53 is an important protein, which has a pivotal role in cell cycle regulation, apoptosis, and angiogenesis. In ATL the operation of TP53 can be influenced by Tax protein as indirectly, downregulation of RPL5 and ATM genes, indirect increase NF-kappa B function, and effect on p38 [10]. As reported before, the reduction of p38 function can happen via downregulation of MAP2K6 and HSPG2 [12]. TP53 also downregulated in TPLL, and sezary syndrome.
ATM is another important protein that downregulated in T-cell malignancy and leads to stimulate the perturbation in DNA repair through the reduction of BRCA1 expression level. In ATL interaction of HTLV-1 p30 with ATM lead to increases viral spread via facilitating cell survival (12).
B. Upregulated hub genes: NTRK1, Neurotrophic Tyrosine Kinase Receptor Type 1, is membrane receptor tyrosine kinase, when bind to its ligand, phosphorylates itself and members of the MAPK pathway. This provides an instructor that is essential for nerve cell development and survival [19]. Upregulation of NTRK1 that happened in TPLL, ATL, and sezary syndrome result in more T-cell proliferation and survival. In addition, activation of the MAPK pathway, induce activation of NFAT and NF-kB therefor stimulate inflammation.
Upregulation of BCL2 and BCL9 occurred in these T-cell malignancies and result in enhancing the survival of malignant cell and activation of Wnt/B catenin.
A membrane protein that highly expressed in the surface of malignant T-cell is DPPIV. This protein has an important function like immune regulation, signal transduction, and apoptosis. CD26/DPPIV plays an important role in tumor biology and is useful as a marker for various cancers [7].
Both LYN and FYN are members of Src kinase, which express in T-cell and neuronal signaling and defined as a proto-oncogene. Deregulation of these signaling is highlighted information of several cancers. FYN encoded a membrane-associated tyrosine kinase that regulates cell growth. FYN induce activation of Janus kinase (JAK), Abl, Src and focal adhesion kinase. LYN activation triggers a cascade of signaling events mediated by Lyn phosphorylation of tyrosine residues within the immune receptor tyrosine-based activation motifs (ITAM) of the receptor proteins. Deregulation of FYN and LYN happened in TPLL and sezary syndrome [20].
In conclusion, a significant aspect of our analyses is the expression alteration of genes accompanied by the inflammation and immune response in ATLL, T-PLL and sezary syndrome developments. Probably, these events are possible strategies that are used in the T-cell malignancies pathogenesis. Eventually, the comprehensive researches based on the low- and high-throughput evaluations are required to accelerate our finding of the complex diseases.
Funding was provided by Mashhad University of medical sciences
- Sud A, Dearden C (2017) T-cell Prolymphocytic Leukemia. Hematol Oncol Clin North Am. 31: 273-283. Link: https://tinyurl.com/y4t9mvoq
- Kakkar N, Calton N, John MJ (2018) T Cell Prolymphocytic Leukemia (T-PLL): Cutaneous Involvement in an Uncommon Leukemia. Indian J Hematol Blood Transfus 34: 573–574. Link: https://tinyurl.com/y3qgdufa
- Nakagawa Y, Hamada T, Matsuda M, Kanno T, Kondo T, et al. (2018) Papuloerythroderma-like cutaneous involvement of a CD62L − subclone of T-cell prolymphocytic leukemia. J Dermatol. Link: https://tinyurl.com/yxntj9kd
- Kotrova M, Novakova M, Oberbeck S, Mayer P, Schrader A, et al. (2018) Next-generation amplicon TRB locus sequencing can overcome limitations of flow-cytometric Vβ expression analysis and confirms clonality in all T-cell prolymphocytic leukemia cases. Cytom Part A 93: 1118–1124. Link: https://tinyurl.com/y5hv4k4x
- Scarisbrick JJ (2018) Infections in mycosis fungoides and Sézary syndrome are a frequent cause of morbidity and contribute to mortality. What can be done? Br J Dermatol 179: 1243–1244. Link: https://tinyurl.com/y2flxuve
- Larocca C, Kupper T (2019) Mycosis Fungoides and Sézary Syndrome. Hematol Oncol Clin North Am 33: 103–120. Link: https://tinyurl.com/yyycal4w
- Nagler AR, Samimi S, Schaffer A, Vittorio CC, Kim EJ, et al. (2012) Peripheral blood findings in erythrodermic patients: Importance for the differential diagnosis of S?zary syndrome. J Am Acad Dermatol 66: 503–508. Link: https://tinyurl.com/y3fot58t
- Ichikawa T, Nakahata S, Fujii M, Iha H, Morishita K (20165) Loss of NDRG2 enhanced activation of the NF-κB pathway by PTEN and NIK phosphorylation for ATL and other cancer development. Sci Rep 5: 12841. Link: https://tinyurl.com/y6smzw7w
- Takasaki Y, Iwanaga M, Tsukasaki K, Kusano M, Sugahara K, et al. (2007) Impact of visceral involvements and blood cell count abnormalities on survival in adult T-cell leukemia/lymphoma (ATLL). Leuk Res 31: 751–757. Link: https://tinyurl.com/y5syy6au
- Jeong SJ, Pise-Masison CA, Radonovich MF, Park HU, Brady JN (2005) Activated AKT regulates NF-κB activation, p53 inhibition and cell survival in HTLV-1-transformed cells. Oncogene 24: 6719–6728. Link: https://tinyurl.com/yy78d884
- Tsuchiya T, Ohshima K, Karube K, Yamaguchi T, Suefuji H, et al. (2004) Th1, Th2, and activated T-cell marker and clinical prognosis in peripheral T-cell lymphoma, unspecified: comparison with AILD, ALCL, lymphoblastic lymphoma, and ATLL. Blood 103: 236–241. Link: https://tinyurl.com/yyyfdp7j
- Mozhgani S-H, Zarei-Ghobadi M, Teymoori-Rad M, Mokhtari-Azad T, Mirzaie M, et al. (2018) Human T-lymphotropic virus 1 (HTLV-1) pathogenesis: A systems virology study. J Cell Biochem 119: 3968–39679. Link: https://tinyurl.com/yxkl5r9v
- Schrader A, Crispatzu G, Oberbeck S, Mayer P, Pützer S, et al. (2018) Actionable perturbations of damage responses by TCL1/ATM and epigenetic lesions form the basis of T-PLL. Nat Commun 9: 697. Link: https://tinyurl.com/y6q8wv28
- Pierobon M, Ramos C, Wong S, Hodge KA, Aldrich J, et al. (2017) Enrichment of PI3K-AKT–mTOR Pathway Activation in Hepatic Metastases from Breast Cancer. Clin Cancer Res 23: 4919–4928. Link: https://tinyurl.com/y4d5sw5q
- Ramakrishnan V, Kumar S (2018) PI3K/AKT/mTOR pathway in multiple myeloma: from basic biology to clinical promise. Leuk Lymphoma 11: 1–11. Link: https://tinyurl.com/yxua4s54
- Keane NA, Glavey SV, Krawczyk J, Dwyer MO (2014) Review AKT as a therapeutic target in multiple myeloma. 897–915. Link: https://tinyurl.com/y6syzwsx
- Harvey RD (2007) PI3 kinase / AKT pathway as a therapeutic target in multiple myeloma. 3: 639–647. Link: https://tinyurl.com/yyc82mjh
- Stoppa G, Rumiato E, Saggioro D (2018) Ras signaling contributes to survival of human T-cell leukemia/lymphoma virus type 1 (HTLV-1) Tax-positive T-cells. Apoptosis 17: 219–228. Link: https://tinyurl.com/y4ceb98n
- Yee AJ, Hari P, Marcheselli R, Mahin- AK, Cirstea DD, et al. (2014) Outcomes in patients with relapsed or refractory multiple myeloma in a phase I study of everolimus in combination with lenalidomide. Br J Haematol 166: 401-409. Link: https://tinyurl.com/y3g8f88n
- Weng Z, Thomas SM, Rickles RJ, Taylor JA, Brauer AW, et al. (1994) Identification of Src, Fyn, and Lyn SH3-binding proteins: implications for a function of SH3 domains. Mol Cell Biol 14: 4509–4521. Link: https://tinyurl.com/y5mg7jvd
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
