Archives of Pulmonology and Respiratory Care
Genotyping and Molecular Characterization of Dermatophytes Isolates Collected from Clinical Samples
Sheikhi Nasrin1, Zaker bostanabad saeed2*, Mirzaahmadi Sina3 and Naziri sahar4
2Department of Microbiology and Biology, Parand branch, Islamic Azad University, Tehran, Iran
3Department of Science and Medicine, Islamic Azad University, Zanjan, Iran
4Mycobacteriology Unit, Masoud Laboratory, Tehran, Iran
Cite this as
Nasrin S, Saeed ZB, Sina M, Sahar N (2017) Genotyping and Molecular Characterization of Dermatophytes Isolates Collected from Clinical Samples. Arch Pulmonol Respir Care 3(1): 052-057. DOI: 10.17352/aprc.000026Background: Detection of dermatophytes by microbiological method is sometimes problematic and some atypical microscopic or macroscopic morphology are non-detectable. Due to morphological similarity and existing intermediate forms and variants, unequivocally separating these dermatophytes is not always straightforward, and sampling appropriate isolates for research is often troublesome. The aim of this study was to compare and evaluate use of sequencing chitin synthase 1 gene (CHS1) with conventional methods for identification of dermatophytes species and we researched the genetic patterns of samples collected for general phylogenetic analysis.
Material and Method: In the primary screening of 250 clinical samples by KOH microscopy method, 64 isolates has been detected as dermatophytes. All samples were cultured and amplified by PCR Method and positive PCR samples have been sequenced. Clinical isolates (64/250) were analyzed by using sequencing gene CHS1 and genotyped by program DNAMAN and MEGA.
Result: The all data were compared with the international database of national center for biotechnology information website. Based on reference sequences of different genotype strains, it was noted that most strains of Trichophyton mentagrophytes were misidentifications of Trichophyton interdigitale.
Conclusion: This research demonstrated that nested PCR and sequencing can be considered as standard method for the diagnosis of dermatophytosis. Also research gives a first result on genetic evolution of the Dermatophytes strains distributing in Iran. It may aid in the creation of a national database that will be a valuable support for further studies.
Introduction
Dermatophytes are keratinophilic fungi capable of causing dermatophytosis and are among the most adaptable parasitic associates of humans. They are classified in three anamorphic genera: Epidermophyton, Microsporum and Trichophyton [1]. Dermatophytes are responsible for most superficial fungal infections and the estimated lifetime risk of acquiring a dermatophyte infection is between 10–20% [2].
Trichophyton rubrum mostly causes mild tinea pedis and onychomycosis. Its transmission from human to human has particularly been promoted by the general use of closed foot wear in urban populations. In older literature [3], the species has been hypothesized to originate from the Far East and subsequently to have been spread by soldiers during the First World War.
In contrast, the T. violaceum complex seems to have gone through speciation processes in Africa and the Middle East. The species comprises some variants or sister species which all mainly cause inflammatory tinea capitis or tinea corporis but differ slightly in cultural characteristics and production of extracellular metabolites: T. gourvilii, T. soudanense, and T. yaoundei [4].
Numerous studies have been conducted on pathogenic dermatophytes in different regions of Iran, and most of these studies have used morphological based criteria which could not reflect the entire spectrum of dermatophytes species [5]. All in all, epidemiology of dermatophytosis in Iran has shown significant changes in various regions of the country over the last decades which is not clarified, is this variation caused by inefficiency of conventional methods or is reflecting the local fungal infection distribution’s pattern [5]. Since the accurate identification of etiologic agents from suspected lesions is important for appropriate treatment and control of potential environmental sources of infection [5], application of conventional methods simultaneously with molecular methods and comparing them considering all involved factors, can lead us to useful and practical information about best identification formula.
Despite large phenetic differences, Trichophyton rubrum and T. violaceum are closely related dermatophytes, composing a single, robust clade in ribosomal DNA (rDNA) internal transcribed spacer (ITS) phylogeny [6].
Some polymorphisms among clinical isolates have been detected in the copy number of a repetitive element (TRS) in the non-transcribed spacer (NTS) of the ribosomal operon, but no clear correlation with either clinical picture or geographic origin was apparent. Detection of additional markers therefore remains necessary [7].
Routine procedures for dermatophytes species identification rely on examination of the colony including; pigmentation of the surface and reverse sides, topography, texture, and rate of growth and also microscopic morphology including; size and shape of macroconidia and micro conidia, spirals, nodular organs, and pectinate branches [8]. Further identification characteristics include nutritional requirements (vitamins and amino acids) and temperature tolerance, as well as urease production, alkaline production of bromocresol purple medium, in vitro hair perforation, etc,, [8,9]. Morphological and physiological characteristics can frequently vary; generally, the phenotypic features can be easily influenced by outside factors such as temperature variation, medium, and chemotherapy [10] and therefore strain identification is often difficult. In the last few years genotypic approaches have proven to be useful for solving taxonomic problems regarding dermatophytes; in fact, genotypic differences are considered more stable and more precise than phenotypic characteristics [8,10].
Molecular methods, such as restriction fragment length polymorphism analysis of mitochondrial DNA[11], sequencing of the internal transcribed spacer (ITS) region of the ribosomal DNA [12], sequencing of protein-encoding genes, arbitrarily primed PCR [AP-PCR] [13], pan-dermatophyte nested PCR and PCR fingerprinting [14], have brought important progress in distinguishing between species and strains.
The genes most frequently used as the main targets for dermatophyte PCR are: the Chitin Synthase 1 (CHS1) gene [15], the ITS regions in the rDNA gene [16] and the topoisomerase II gene [17]. In the last couple of years, the field of fungal characterizations by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF) has been explored [18]. PCR technology is simple, rapid and, in the absence of specific nucleotide sequence information for the many dermatophyte species, able to generate species- specific or strain-specific DNA polymorphisms on the basis of characteristic band patterns detected by agarose gel electrophoresis [13]. This provides significant possibilities for diagnostic and epidemiological typing studies.
The aim of this study was to identification and comparing PCR by using primers targeting the dermatophyte-specific sequence of the CHS1 gene with culture isolation for diagnosis of 250 clinically suspected received by our diagnostic laboratory in a six- month period from August 2014 to January 2015. Also in this study, we have tried a general phylogenetic analysis for all samples by single fragment.
Materials and Methods
Fungal strains. In all, 250 clinical and reference strains from several geographic regions were collected in Massoud laboratory and Razi hospital, Tehran, Iran during 6-month period from August 2014 and January 2015 and identified using smear, conventional culture and molecular techniques and causing different clinical pictures analyzed. For DNA extraction, strains were cultured on sabouraud dextrose agar [4]. Morphological identification was done at the host laboratories.
A. KOH microscopy
Samples were collected and transported in a sterile black paper, to avoid exposure to moisture and prevent the growth of contaminants. Most mycological specimens should be examined in a fluid state, usually after partial digestion with potassium hydroxide. Place epidermal scales on a microscope slide cover with 10% KOH and a cover slip warm, warm bellow the boiling point over a small flame and examine under the microscope Giemsa stain and gram & ziehl-neelsen stains of dried smear is useful in diagnosis [19].
B. Culture conditions
In general specimens are taken for culture as directed above for microscopic examination. Specimens should be planted on agar media (modified sabouraud dextrose agar with antibiotics). Incubate slants at temperature 25-30C and should be examined at intervals during incubation so that if they are contaminated the pathogen can be transferred to new slant before being over grown with the contaminant.
In general it’s best to examine a young culture when it first begins to form spores & again a few days later. Slide culture of various types used for the microscopic examination of fungus. It often is more convenient to examine mycelium directly from an agar plate [19].
C. Purification of fungal genomic DNA and PCR
DNA extraction was performed on the portion of specimen by using a NA DNA technology kits. First-round PCR was performed using primer pairs CHS1 1S (5’-CAT CGA GTA CAT GTG CTC GC-3’; nucleotides [nt] 70 to 89) and CHS1 1R (5’-CTC GAG GTC AAA AGC ACG CC-3’; nt 485 to 504). These primers amplify a 435-bp DNA fragment of the dermatophytes-specific CHS1 gene sequence of Arthroderma benhaemiae, a teleomorph of Trichophyton mentagrophytes (DDBJ accession no. AB003558) [20]. Nested PCR was performed by designing a novel set of primers, JF2 (5’-GCA AAG AAG CCT GGA AGA AG-3’; nt 111 to 130) and JR2 (5’-GGA GAC CAT CTG TGA GAG TTG-3’; nt 378 to 398), amplifying a DNA fragment of 288 bp from the internal sequence of the amplicon obtained from first-round PCR. The PCR mixture (25 μl) for first-round PCR contained 12.5 μl of 2x Master Mix (Amiplicon III, Viragen, Mgcl2 2.0 mM), 25 pmol each of primers CHS1 1S and CHS1 1R (TAG Copenhagen A/S), and 5 μl of DNA template. Deionised water was added subsequently to achieve the reaction volume. The reaction mixture was initially denatured at 94°C for 3 min, followed by 30 cycles of denaturation at 94°C for 60 s, annealing at 60°C for 75 s, and extension at 72°C for 120 s. This was followed by a final extension step for 7 min at 72°C in a thermal cycler (PeQ Lab biotechnologie GmbH). The PCR mixture for nested PCR consisted of 25 pmol of primers JF2 and JR2 along with a 1:6 diluted product of the primary cycle as the DNA template; the rest of the constituents were the same as those described above. The running conditions of nested PCR were similar to the first-round PCR except that an annealing temperature of 63°C and 40 cycles were used. Triple-distilled water and DNA of Trichophyton mentagrophytes were used as the negative and positive controls, respectively.
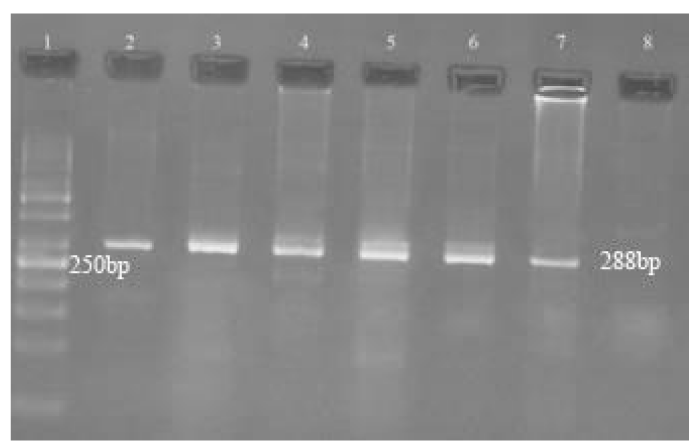
To document the amplified products, 5 μl of product from nested PCR was electrophoresed on a 1.5% agarose gel (containing Safe View DNA Stain) in Tris-borate-EDTA buffer, initially at 100 V for 5 min and then at 80 V for 60 min. Thereafter, bands were visualized under UV light and amplicon of 288 bp was taken as positive for dermatophytes (Figure 1). Based on the result of culture identification some of the positive PCR products were sequenced and analysed by online software.
Results
KOH microscopy and Culture
The age of the study population ranged between 11 and 87 years. Of the 250 clinically suspected cases of skin dermatophytosis, 25.6% (64/250) were positive for fungal elements by KOH microscopy that contain 41 males and 23 females. Positive dermatophytes samples by KOH microscopy were culured and 62.5 %( 40/64) cases isolated. Among the dermatophytes isolated on culture Trichophyton mentagrophyte was the commonest isolate (48%, 16/40), followed by Epidermophiton. Flocosome (15%, 6/40), Microsporom canis (12.5%, 5/40), Trichophyton vorecosom (12.5%, 5/40), Trichophyton rubrum (12.5%, 5/40), Trichophyton tonsurans (5%, 2/40) and 1(2.5%) case was not identified. The remaining 24 specimens, 16 cases had no qrowth on culture, 2 specimens were positive for nondermatophytic molds and 6 cases were contaminationed.
PCR and DNA sequencing analysis
Among 64 specimens, 62 (96.8%) cases were detected by nested PCR as Dermatophytes. Morphological similar isolates obtained from culture were placed in the same group. Some samples from each group was selected to study the gene sequence and also the samples were not identified or suspected by culture were evaluated by sequencing (Table 1). Sequences of the isolates entered in software JPhydit, compared with the corresponding sequence database and species identification was made. Of the 62 nested PCR positive specimens, candida albicans was cultured from 1 specimen, thus nested PCR detecting cases with hidden mixed infections.
Of 9 identified with the culture as a Trichophyton mentagrophytes, 8 cases Trichophyton Interdigital (varieties Trichophyton) and 1 case (1/11%), Trichophyton Equinum was identified by sequencing.Of 4 pidermophyton floccosum identified with the culture, 3 (75%) of them were confirmed by sequencing and 1 (25%) was identified Trichophyton Interdigital. From 5 T.verrucosum identified with culture, only 1 patient (20%) were confirmed by sequencing, 2 (40%) Epidermophyton floccosum and 2 patients (40%), Trichophyton Interdigital was obtained by sequencing.4 identified with the culture of Trichophyton rubrum, 2 (50%) were confirmed by sequencing and 2 patients (50%) Trichophyton Interdigital, were identified.Of 1 Trichophyton Tunsrans identified with the culture, Trichophyton Interdigital were identified by sequencing.3 identified with the culture of M. canis, 2 (6/66%) were confirmed by sequencing and 1 (3/33%) were identified with sequencing as a Trichophyton mentagrophytes.
Discussion
The low success rate of PCR primers derived from the library suggested that the majority of the clones were chimeric and not representative of contiguous stretches of genomic DNA. To reduce the number of recombinant clones, it may be necessary to run fewer cycles of PCR amplifications before and after the enrichment procedure [4].
Dermatophytes are among the few fungi causing communicable diseases. However previously most dermatophyte strains had relatively restricted geographical distribution, but recently, dermatophytosis has become one of the most common human infectious diseases in the world and is cosmopolitan in distribution.
The striking phenetic deviations from T. violaceum despite uniformity at the molecular level indicate that speciation processes in dermatophytes are of recent date. However, no clear association was found with the clinical picture or the geographic origin.
Dermatophytosis cannot be easily diagnosed on the basis of clinical manifestations [21]. Further it is more difficult to diagnose dermatophytosis in immunocompromised patients; since clinical presentation is often atypical [20]. The differential diagnosis of dermatophytoses includes seborrhoeic dermatitis, atopic dermatitis, contact dermatitis, psoriasis, candidal intertrigo, erythrasma, Eczema etc [21]. We have not demonstrated innovation and invention samples in this study and only have isolated species from different region of Tehran city that is capital of Iran. Despite the multicopy gene structure of the NTS region, it was suggested by Yazdanparast et al. [20], that multiple strains can be involved in an infection of a single patient.
Good laboratory methods are available for rapid and accurate identification of the dermatophytes involved, in order to apply appropriate treatment and prevention measures. The conventional methods of fungal detection have their own drawbacks; for e.g. KOH microscopy has low specificity and fungal culture can identify specific pathogen but it takes a long incubation period [21]. The false negative rate of fungal culture is approximately 30% and the sensitivity was about 60% [20]. Most of the dermatophyte isolates from patients on antifungal treatment generally do not show characteristic morphology on culture, thus cause to confuse the results of culture isolation [22]. The changing profiles of human dermatophytoses among countries have further necessitated the development of improved diagnostic methods for identification of dermatophytes [22]. Therefor newer fungal diagnostic methods are required for identification of the etiological agent, not only for accurate diagnosis, but also for post-therapeutic strategies [23,24]. In our study; Frequency of dermatophytosis was more prevalent in men (64.0%) than women (36.0%). The high prevalence in males has been reported in several reports [25-27]. It may be due to occupational contact in males as they more involved in outdoor activities.
In the present study, the number of positive samples for fungi by culture on SCC were 40 (62.5%) samples which is near to that detected by Lopes et al. [28], (56.6%), Pontes et al. [29], (66.5%), and El- Batawi et al. [30], (68.7%).Trichophyton mentagrophyte was the commonest isolate (48%, 16/40), followed by Epidermophiton. Flocosome (15%, 6/40), Microsporom canis (12.5%, 5/40), Trichophyton vorecosom (12.5%, 5/40), Trichophyton rubrum (12.5%, 5/40), Trichophyton tonsurans (5%, 2/40) and 1(2.5%) case was not identified. The remaining 24 specimens, 16 cases had no qrowth on culture, 2 specimens were positive for nondermatophytic molds and 6 cases had contamination.
T. rubrum, as now recircumscribed on the basis of microsatellite data, occurs in Canada and Japan with polymorphic populations (A/B). Polymorphism is noted on a very small geographic scale, since several cities in Japan, as well as Toronto in Canada, harbor both genotypes. In Japan genotypes A and B are nearly equally frequent (45% versus 55%). Rippon [14], suggested that T. rubrum has evolved from a chronic case of tinea corporis in the late 19th century in an area of endemicity in Southeast Asia [4].
In the past few years several molecular methods for the detection and identification of dermatophytes from clinical samples have been developed. Major difficulties of PCR methods are that it requires training, sophisticated equipments and standardization and it is also expensive [31]. On the other hand, it is not only sensitive and specific, but also has the potential to decrease the time taken for the laboratory identification of pathogens that grow slowly or are difficult to culture. By the use of PCR, reliable rapid results within 24 hours in contrast to the 21 days of incubation required for the isolation of dermatophytes by culture. The application of PCR technology directly to the clinical specimens would allow early and accurate identification of agents of dermatophytosis. This would permit prompt and targeted initiation of antifungal therapy [32]. Chi-square test results to determine the relationship between culture and sequencing represents a significant difference between the two diagnostic methods (CI = 0.95; P = 0.000).
Uniformity in T. rubrum populations (An in Africa and B in the United States and Europe) would suggest either a recent emergence of genotypes favored by natural selection of, e.g., virulent strains, or of recent bottlenecks in population size provided that our nonrandom sampling is representative for each of the continents. In clonal (no recombining) organisms we would expect to have similar genetic population signatures for both situations. In the case of a recently emerged pathogen, however, we would expect no or low geographical structuring combined with an absence of host specificity of the genotypes [32], since the species is supposed to have a low degree of adaptation to any new host. T. rubrum is known to have a long standing relationship with the human host and is only rarely encountered in animals.
It should be noted, that for special analysis phylogenetic by MLSA (multi locus sequence analysis) must be do sequence ITS, rDNA and mDNA genes and regions for evaluating result.
According to Chi-square test which is based on the location of the infection, there is no significant difference in terms of type of dermatophytes (the growth of dermatophytes in skin all over the body surface is provided).It may therefore be concluded that nested PCR and seqencing targeting the CHS1 gene may be considered the gold standard for detection of dermatophytes in patients and can aid the clinician in initiating prompt and appropriate antifungal therapy. This technique may also play an important role in large-scale studies and in the management of problematic cases of dermatophytosis.
- Li HC, Bouchara JP, Hsu MM, Barton R, Su S, et al. (2008) Identification of dermatophytes by sequence analysis of the rRNA gene internal transcribed spacer regions. J Med Microbiol 57: 592–600. Link: https://goo.gl/S8b4yq
- Jaya Garg, Ragini Tilak, Atul Garg, Pradyot Prakash, Anil Kumar, et al. (2009) Rapid detection of dermatophytes from skin and hair. BMC Research Notes 2: 60. Link: https://goo.gl/ukU2Yc
- Gräser Y, Kühnisch J, Presber W (1999) Molecular markers revealexclusively clonal reproduction in Trichophyton rubrum. J Clin Microbiol 37: 3713–3717. Link: https://goo.gl/DqxOuC
- Ohst T, de Hoog S, Presber W, Stavrakieva V, Gräser Y (2004) Origins of Microsatellite Diversity in the Trichophyton rubrumT. violaceum Clade (Dermatophytes) J Clin Microbiol 42: 4444–4448. Link: https://goo.gl/5oCius
- Ghojoghi A, Falahati M, Pagheh AS, Abastabar M, Ghasemi Z (2015) Molecular Identification and Epidemiological Aspects of Dermatophytosis in Tehran Iran. Res Mol Med 3: 11-16. Link: https://goo.gl/PcnG66
- Gräser Y, Kuijpers AF, Presber W, de Hoog GS (2000) Molecular taxonomy of the Trichophyton rubrum complex. J Clin Microbiol 38: 3329-3336. Link: https://goo.gl/UISdBb
- Jackson CJ, Barton RC, Kelly SL, Evans EG (2000) Strain identification of Trichophyton rubrum by specific amplification of subrepeat elements in the ribosomal DNA nontranscribed spacer. J Clin Microbiol 38: 4527-4534. Link: https://goo.gl/7wDW5F
- Faggi E, Pini G, Campisi E, Bertellini C, Difonzo E, et al. (2001) Application of PCR to Distinguish Common Species of Dermatophytes J Clin Microbiol 39: 3382–3385. Link: https://goo.gl/bk8nb2
- Weitzman I, Summerbell RC (1995) the dermatophytes. Clin Microbiol Rev 8: 240–259. Link: https://goo.gl/VJW4Ld
- Liu D, Coloe S, Baird R, Pedersen J (2000) Application of PCR to the identification of dermatophyte fungi. J Med Microbiol 49: 493–497. Link: https://goo.gl/4x7FJx
- Kawasaki M, Aoki M, Ishizaki H, Nishimura K, Miyaji M (1996) Phylogeny of Epidermophyton floccosum and other dermatophytes. Mycopathologia 134: 121–128. Link: https://goo.gl/u8W3vl
- Mochizuki T, Tanabe H, Kawasaki M, Ishizaki H, Jackson CJ (2003) Rapid identification of Trichophyton tonsurans by PCR-RFLP analysis of ribosomal DNA regions. J Dermatol Sci 32: 25-32. Link: https://goo.gl/mS58jT
- Liu, D, S Coloe, R Baird, J. Pedersen (2000) Application of PCR to the identification of dermatophyte fungi. J Med Microbiol 49: 493–497. Link: https://goo.gl/oGbuUj
- Garg J, Tilak R, Singh S, Anil K Gulati, Atul Garg, et al. (2007) Evaluation of pan-dermatophyte nested PCR in diagnosis of onychomycosis. J Clin Microbiol 45: 3443–3445. Link: https://goo.gl/Vu7RvV
- Kano R, Hirai A, Muramatsu M, Watari T, Hasegawa A (2003) Direct detection of dermatophytes in skin samples based on sequence of the chitin synthase 1 (CHS1) gene. J Vet Med Sci 65: 267–270. Link: https://goo.gl/6RrZDk
- Rezaei-Matehkolaei A, Makimura K, Shidfar MR, Eshraghian, N Jalalizand, et al. (2012)Use of single-enzyme PCR restriction digestion barcode targeting the internal transcribed spacers (ITS rDNA) to identify dermatophyte species. Iran J Publ Health 41: 82–94.Link: https://goo.gl/EIpJ2R
- Kanbe T, Suzuki Y, Kamiya A, Mochizuki T, Fujihiro M, et al. (2003) PCR based identification of common dermatophyte species using primer sets specific for the DNA topoisomerase II genes. J Dermatol Sci 32:151-61. Link: https://goo.gl/3YGII0
- Alshawa K, Beretti J-L, Lacroix C, Brunhilde Dauphinc, Gilles Quesnea, et al. (2012) Successful identification of clinical dermatophyte and Neoscytalidium species by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 50: 2277–2281. Link: https://goo.gl/ZIyLDB
- Emmons CW, Binford CH, Utz JP, Kwon-Chung KJ. Medical mycology. New Edition.
- Garg J, Tilak R, Garg A, Prakash P, Gulati AK, et al. (2009) Rapid detection of dermatophytes from skin and hair. BMC Research Notes 2: 60 Link: https://goo.gl/MFkkJl
- Barry I, Hainer BL (2003) Dermatophyte Infections. American family physician 67: 101-108. Link: https://goo.gl/QRCYLt
- Liu, Coloe S, Baird R, Pedersen J (1997) Rapid differentiation of Microspovum dermatophytes based on arbitrarily primed PCR amplification. Opportunistic Pathogens 9: 3-6. Link: https://goo.gl/0Q0uXM
- Shiraki Y, Soda N, Hirose N, Hiruma M (2004) Screening Examination and Management of Dermatophytosis by Trichophyton tonsurans in the Judo Club of a Universitys. Nippon Ishinkin Gakkai Zasshi 45: 7-12. Link: https://goo.gl/pCIKp8
- Urano S, Shirai S, Suzuki Y, Sugaya K, Takigawa M, et al. (2003) A case of Tinea capitis caused by Trichophyton tonsurans. Jpn J Med Mycol 44: 25-29. Link: https://goo.gl/GaN1bE
- Mahmoudabadi AZ (2005) A study of dermatophytosis in South West of Iran (Ahwaz). Mycopathologia 160: 21–24. Link: https://goo.gl/lfKlkL
- Bhatia VK, Sharma PC (2014) Epidemiological studies on Dermatophytosis in human patients in Himachal Pradesh, India. SpringerPlus 3: 134-141. Link: https://goo.gl/L0ELpU
- Rassai S, Feily A, Sina N, Derakhshanmehr F (2011) Some Epidemiological Aspects of Dermatophyte Infections in Southwest Iran. Acta Dermatovenerol Croat 19: 13-15. Link: https://goo.gl/QyWKPN
- Lopes JO, Alves SH, Mari CRD, To L, Brum LM, et al. (1999) A ten year survey of onychomycosis in the central region of the Rio Grande do Sul, Brazil. Rev Inst Med Trop São Paulos 41: 147-149. Link: https://goo.gl/zdilIv
- Pontes ZB, Lima Ede O, Oliveira NM, Dos Santos JP, Ramos AL, et al. (2002) Onychomycosis in João Pessoa city, Brazil. Rev Argent Microbiol 34: 95-99. Link: https://goo.gl/YSfdmM
- El Batawi MM, Arnaot H, Shoeib S, Bosseila M, El Fangary M, et al. (2006) Prevalence of Non- Dermatophyte Molds in Patients with Abnormal Nails. Online Egypt Dermatol J 2: 12. Link: https://goo.gl/EQ2iKv
- Arca E, Saracli MA, Akar A, Yildiran ST, Kurumlu Z, et al. (2004) Polymerase chain reaction in the diagnosis of onychomycosis. Eur J Dermatol 14: 52-55. Link: https://goo.gl/HNRCU0
- Bagyalakshmi R, Senthilvelan B, Therese KL, Murugusundram S, Madhavan HN (2008) Application of polymerase chain reaction (PCR) and PCR based restriction fragment length polymorphism for detection and identification of dermatophytes from dermatological specimens. Indian J Dermatol 53: 15-20. Link: https://goo.gl/YaaqQw
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
