Archives of Hepatitis Research
The dark proteome of rodent hepatitis E virus: Analysis of intrinsically disordered regions
Zoya Shafat1, Anwar Ahmed2, Mohammad K Parvez3, Asimul Islam1 and Shama Parveen1*
2Centre of Excellence in Biotechnology Research, College of Science, King Saud University, Riyadh, Saudi Arabia
3Department of Pharmacognosy, College of Pharmacy, King Saud University, Riyadh, Saudi Arabia
Cite this as
Shafat Z, Ahmed A, Parvez MK, Islam A, Parveen S (2022) The dark proteome of rodent hepatitis E virus: Analysis of intrinsically disordered regions. Arch Hepat Res 8(1): 005-011. DOI: 10.17352/ahr.000032Copyright License
© 2022 Shafat Z, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Hepatitis E virus (HEV) is the causative agent of Hepatitis E infections across the world. Intrinsically disordered protein regions (IDPRs) or Intrinsically Disordered Protein (IDPs) are regions or proteins that are characterized by a lack of definite structure. These regions or proteins play significant roles in a wide range of biological processes, such as cell cycle regulation, control of signaling pathways, etc. IDPRs or IDPs in proteins are associated with the virus’s pathogenicity and infectivity. The occurrence of intrinsic disorder in the proteome of rat HEV remains to be elucidated, which prompted us to explore its dark proteome. In this study, the unstructured/disordered regions of ORF proteins of rat HEV have been examined. We have analyzed the prevalence of intrinsic disorder by using a set of computational predictors. The intrinsic disorder propensity analysis showed that the ORF proteins consisted of a varying fraction of intrinsic disorder. The ORF3 protein was identified with a maximum propensity for intrinsic disorder while the protein ORF6 showed the least propensity for the intrinsic disorder. Further, the analysis revealed ORF6 as highly structured protein (ORDP); ORF1 and ORF4 as moderately disordered proteins (IDPRs); and ORF3 and ORF5 as highly disordered proteins, categorizing them as ordered protein (ORDP), a protein having Intrinsically Disordered Region (IDPR) and Intrinsically Disordered Proteins (IDP) respectively. Such disordered regions may play several important roles in the pathogenesis and replication of viruses. Collectively, this comprehensive study data from our investigation suggested ORF protein’s role in the regulation and pathogenesis of rat herpesvirus.
Background
Hepatitis E is inflammation of the liver which is caused by the Hepatitis E virus (HEV) [1]. Worldwide, about 20 million HEV infections and 3.3 million symptomatic hepatitis E cases occur annually which results in 44,000 deaths [2]. HEV is of the family Hepeviridae and belongs to the genus Orthohepevirus [3]. The genome of HEV is a single-stranded positive-sense RNA (7.2 kb in length), which is flanked with short 5′ and 3’ non-coding regions (NCR) [4]. The HEV genome comprises three open reading frames (ORFs): ORF1, ORF2, and ORF3. The ORF1 codes for the viral non-structural polyprotein (pORF1), the ORF2 codes for the viral capsid protein (pORF2), and ORF3 codes for the viral pleiotropic protein (pORF3) [5].
Besides transmission of HEV through the fecal-oral route (in developing nations), It has been suspected that human infections occur mainly due to zoonotic transmission of HEV that occurs, it is suspected that these human infections result from zoonotic transmission of GT 3 of HEV, wherein, wild boars, domestic pigs and deer act as major reservoirs or host organisms [6]. In particular, commensal rodents also act as an additional reservoir for HEV and may play a major in the epidemiology of hepatitis E [7-9]. Though the genome of rodent herpesviruses is similar in organization to human herpesviruses, however, it has been identified in the year 2009 that the genome of rats from Norway had a dissimilar organization in comparison to other herpesviruses strains [10]. The two complete nucleotide sequences were analyzed from Norway in Germany which suggested a completely separate genotype for these rodent herpesviruses [10]. However, it has been predicted through software that these Norway rat herpesviruses consisted of some additional open reading frames, i.e., ORF1, ORF2, ORF3, ORF4, ORF5, and ORF6 [10]. These nucleotide sequences had high divergence to other HEV strains, i.e., HEV G1, HEV G2, HEV G3, HEV G4, and avian HEV strains [10]. It was also identified that, unlike typical HEV genomic organization, the ORFs ORF1 and ORF3 do not overlap in these two rat HEVs strains [10]. Three additional putative ORFs of 280 - 600 nt that overlap with ORFs 1 or 2 were predicted for each rat HEV genome strain [10]. In this context, this study aims to summarize the features of the ORF encoded proteins of this particular rodent herpesvirus (obtained from Norway Germany).
Recent studies have determined the role of different reading frame encoded proteins in HEV regulation by analyzing their intrinsically disordered regions [11-14], as these regions are linked with virus’s infection and pathogenesis [15-17]. However, a direct correlation between the disordered segments of ORF encoded proteins and viral adaptation has not been discovered in Norway strain comprising additional reading frames. Thus, we attempted to delineate the role of these ORF encoded proteins in rat HEV pathogenesis.
The present study analyzed the structurally “unknown” regions (i.e., a fraction of a proteome that has no detectable similarity to any PDB structure) of the rat HEV. This fraction we call the “dark proteome.” The proteins or protein regions that fail to get folded into definite three-dimensional (3D) structures but remain biologically active are termed as intrinsically disordered proteins (IDPs) and intrinsically disordered protein regions (IDPRs), respectively [18-20]. These disordered protein regions exist as extremely active ensembles that are rapidly interconvertible under different physiological conditions [19-21]. Due to the occurrence of the peculiar phenomenon, i.e., binding of several disordered regions to one ligand or vice-versa (one disordered region binds to many partners), the intrinsically disordered regions are utilized in protein-protein interactions [22,23]. The prevalence of the intrinsic disorder in the proteome of rat herpesvirus remains unknown. The current study reports analysis on the disordered side of rat HEV using computational methods to check the occurrence of intrinsically disordered regions in order to shed some light on their disorder-related functions in HEV adaptation.
Materials and methods
Sequence retrieval
The protein sequence (Accession ID: GU345043) of rat HEV was obtained from the NCBI (National Center for Biotechnology Information) GenBank database.
Structural analysis
The rat HEV proteins three-dimensional (3D) structural models were automatically generated using Phyre2 (Protein Homology/AnalogY Recognition Engine) server (http://www.sbg.bio.ic.ac.uk/~phyre2/html/page.cgi?id=index) and were used for comparative analysis.
Intrinsic disorder prediction
9Intrinsically disordered regions (IDRs) of the rat HEV proteome were predicted using the PONDR® (Predictor of Natural Disordered Regions) (www.pondr.com) at its default settings. Multiple predictors such as members of the PONDR® family including PONDR®VLS2, PONDR®VL3, and PONDR® VLXT were exploited to predict the intrinsic disorder predisposition in rat HEVs. This bioinformatics tool predicts the residues or regions which fail in the propensity for an ordered structure formation. The protein residues with predicted scores between 0.2 and 0.5 were considered as flexible, while the residues which had scores, exceeding the 0.5 threshold value, were predicted as intrinsically disordered ones.
Results
Analysis of structure
The 3D modeled structures of the ORF proteins of rat HEV were generated through the Phyre2 web server as shown in Figure 1A–F.
The predicted percentage of alpha-helix, beta-strand, and disordered residues in the generated 3D rat HEV proteins are summarized in Table 1.
Therefore, the initial structural analysis revealed that all the rat HEV proteins consisted of disordered regions (Figure 1A-F).
Analysis of protein intrinsic disorder predisposition
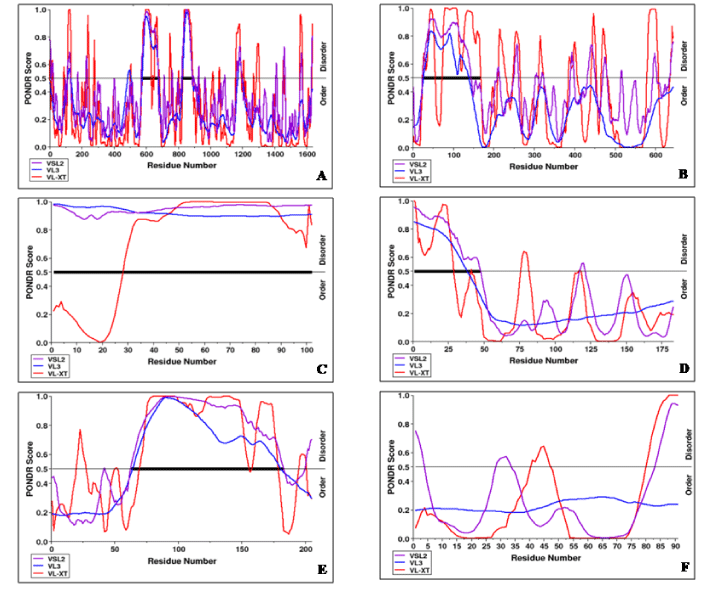
The intrinsic disorder propensity analysis of rat HEV proteins was carried out to elucidate their intrinsic disorder properties. The predicted intrinsic disordered residues obtained from three disorder predictors for ORF encoded proteins of rat HEV are mentioned in Table 2. The resulting disorder profiles of the rat HEV proteins are shown in Figure 2A-F.
On the basis of the predicted percentage of intrinsic disorder and the presence of disordered domain, the different ORF proteins of rat HEV were categorized as follows:
ORF1 protein: The intrinsic disorder analysis showed ORF1 protein as a moderately disordered protein, as it consisted of less than 30% (VLXT, VSL2, and VL3) of the disordered residues in its polypeptide chain with two stretches of disordered domains at positions distinct from N- and C-terminals. Thus, it was categorized into IDPRs, i.e., structured proteins with intrinsically disordered segments of proteins possessing both structured unstructured regions (Table 2).
ORF2 protein: The intrinsic disorder analysis showed ORF2 protein as a highly disordered protein, as it consisted of >30% (as predicted by VLXT and VSL2) and moderately disordered as it consisted of less than 30% (as predicted by VL3) of the disordered residues in its polypeptide chain along with the presence of disordered domain. Thus, on combining these assumptions it was categorized into both IDPs, i.e., proteins having a significant fraction of disordered regions, or IDPRs, i.e., structured proteins with intrinsically disordered segments (Table 2).
ORF3 protein: The intrinsic disorder analysis showed ORF3 protein as a highly disordered protein, as it consisted of >30% (VLXT, VSL2, and VL3) of the disordered residues in its polypeptide chain. Thus, it was categorized into IDPs (Table 2).
ORF4 protein: The intrinsic disorder analysis showed ORF4 protein as a moderately disordered protein, as it consisted of less than 30% (VLXT, VSL2, and VL3) of the disordered residues in its polypeptide chain with a stretch of a disordered domain at the N-terminus. Thus, it was categorized into IDPRs, i.e., structured proteins with intrinsically disordered segments (Table 2).
ORF5 protein: The intrinsic disorder analysis showed ORF5 protein as a highly disordered protein, as it consisted of >30% (VLXT, VSL2, and VL3) of the disordered residues in its polypeptide chain along with possession of long disordered domain towards the C-terminus. Thus, it was categorized into IDPs (Table 2).
ORF6 protein: The intrinsic disorder analysis showed ORF6 protein as a structured protein, as it consisted of less than 30% (VLXT, VSL2, and VL3) of the disordered residues in its polypeptide chain without the presence of any disordered domain. Thus, it was categorized into ORDPs, i.e., proteins possessing a significant amount of structure (Table 2).
Discussion
The intrinsic disorder is linked with the pathogenesis and infection of the viruses [15-17]. To complete the life cycle, viruses require various interactions with the components of the host cells. Beginning from the virus’s attachment, its entry, commandeering the host machinery, synthesis of the viral components, and particle assembly to the last phase, i.e., exiting as new infectious particles from the host cell [24]. All these stages rely heavily on the intrinsic disorder prevalent in viral proteins [24]. The biology of the unstructured regions of the Norway rat HEV, comprising additional reading frames, remains to be explored. Therefore, the present study reports the analysis on the unstructured regions of the ORF encoded proteins of rat herpesvirus to shed novel light on its functionality in HEV regulation.
Analysis of protein structure provides a detailed understanding of its function. In this context, the rat HEV protein structures were examined using a web portal for protein modeling and analysis. A study has suggested that loops/coils are not necessarily disordered, however, protein disorder is only found within loops [25]. Thus, it was revealed that the modeled 3D structures of rat HEV proteins were identified with all three major secondary structure states, i.e., alpha-helix, beta-strand, and loops/coils. Therefore, our initial investigation showed the prevalence of the intrinsic disorder in the rat HEV proteome. The specific role of disordered regions in several nonstructural proteins has been demonstrated to participate in the multiplication and regulatory functions of viruses [26]. For instance, a recent study has shown the involvement of ORF4 protein in the regulation and pathogenesis of HEV due to the presence of a significant fraction of disordered regions [12]. The disordered regions in the ORF1 Y-domain of HEV have also been shown to perform a crucial role in its pathogenesis due to its intrinsic disorder phenomenon [11]. In HDV (hepatitis delta virus), the translation of a delta antigen (a single basic protein) forms the basis of its replication, which is considered as an IDP molecule [27]. Via both experimental and computational studies [28]. The HCV (Hepatitis C virus) interacts with several viral and host proteins required for its replication via its disordered nonstructural NS5A protein domain [29,30]. These protein-protein interactions result in the occurrence of several significant biological functions. Moreover, the PPR (Polyproline region) of nonstructural ORF1 has been associated with the regulation of HEV in addition to its role in replication, due to its characteristic intrinsic disorder property [31].
The rat HEV proteins were initially categorized on the basis of the overall degree of intrinsic disorder. The categories included structured proteins (0–10%), moderately disordered proteins, and highly disordered proteins (30–100%) [32,33]. Additionally, the ORF proteins were categorized on the basis of the length of disordered domains and an overall fraction of disordered residues. The categories consisted of ordered proteins (ORDPs); intrinsically disordered protein regions (IDPRs) and intrinsically disordered proteins (IDPs) [18,32]. ORDPs are intrinsic disorder protein variants that consist of less than 30% of disordered residues without disordered domain (consecutive disordered residues) at either C- or N-terminus or in positions distinct from the N- and C-terminals. IDPRs are intrinsic disorder protein variants that consist of less than 30% of disordered residues with a disordered domain at either C- or N-terminus or in positions distinct from the N- and C-terminals. IDPs are intrinsic disorder protein variants that consist of more than 30% of disordered residues. On summing up these criteria, our intrinsic disorder propensity analysis revealed ORF3 protein as the most disordered protein and ORF6 protein as the most ordered protein in the rat HEV proteome. Interestingly, we found out that the rat HEV proteome was identified with all the three intrinsic disorder variants, such as ORDP, IDPR, and IDP (Table 3).
Recent investigation on HEV proteome, consisting of three ORF encoded proteins (ORF1, ORF2, and ORF3), was carried out by analyzing its intrinsic disorder [14]. The [14]. In the current study, ORF3 protein possessed the highest fraction of intrinsic disorder suggesting it as an IDP which shows consistency with the previous study suggesting ORF3 as a highly disordered protein (IDP) [14]. Moreover, our result on ORF2 protein substantiates the previous finding which showed ORF2 protein possessed a disordered segment at its N-terminus [14]. Furthermore, our analysis revealed ORF4 as an IDPR which is in line with the previous study that showed the ORF4 obtained from the host rat is an IDPR variant [12]. Taken together our observations, could be interpreted that ORF3 has the highest percentage of a fraction of disordered residues followed by ORF2 and ORF1 has the least fraction of disordered residues. Thus, it is noteworthy to mention that our findings are in agreement with the earlier study which demonstrated that the ORF3 had the highest prevalence of disordered residues followed by ORF2, which had a comparatively lesser fraction of intrinsic disorder, while the ORF1 had the least number of disordered residues in the HEV proteome [14].
The “IDPR/IDP” is defined as the disordered region in protein or disordered protein. These regions/proteins perform several significant roles in a variety of biological processes, such as control of signaling pathways, cell cycle regulation, etc. [16,22,23]. It has been suggested that IDPRs/IDPs achieve their signaling cascade by binding to their partners with low affinity and high specificity [34]. Thus, the proteins, such as ORF1, ORF2, and ORF4 can play crucial roles in important biological processes as IDPRs. IDP plays a significant role in the recognition, signaling, regulation, and control of Protein-Protein Interaction (PPI) networks [35]. IDPs form essential components of cellular signaling machinery due to their ability to interact differently which results in different consequences [36]. Moreover, they are characterized by enormous flexibility and random conformation (coiled-like). Thus, taken together, these distinctive features enable IDPs to participate in one too many and vice-versa interaction [37-39]. Particularly, like IDP, ORF3 may possibly perform a crucial role in the viral regulation via PPI.
Thus, taken together, our analysis suggests that the disordered regions prevalent in rat HEV proteome, as IDPRs/IDPs, could perform significant and diverse biological roles through PPIs.
Conclusion
The current study provides novel intrinsic disorder analysis on the rat HEV proteome. Our data revealed the occurrence of all intrinsic disorder variants (ORDP, IDPR, and IDP) in the proteome. The ORF3 protein was identified as the most disordered protein and ORF6 protein consisted of the least fraction of intrinsic disorder. The occurrence of the unstructured regions suggested that rat HEV proteins could be engaged in diverse and essential biological functions. Further, complete experimental insights into the disorders of these viral proteins might help in identifying protein functions and the biology of rat HEV.
The authors would like to acknowledge Maulana Azad National Fellowship (MANF), University Grant Commission (UGC), Council of Scientific and Industrial Research (CSIR) (37(1697)17/EMR-II) and Central Council for Research in Unani Medicine (CCRUM), Ministry of Ayurveda, Yoga and Neuropathy, Unani, Siddha and Homeopathy (AYUSH) (F.No.3-63/2019- CCRUM/Tech) supported by the Government of India.
- Alves C, Cheng H, Roder H, Taylor J (2010) Intrinsic disorder and oligomerization of the hepatitis delta virus antigen. Virology 407: 333–340. Link: https://bit.ly/3t6bhTd
- Arankalle VA, Joshi MV, Kulkarni AM, Gandhe SS, Chobe LP, et al. (2000) Prevalence of anti‐hepatitis E virus antibodies in different Indian animal species. J Viral Hepat 8: 223-227. Link: https://bit.ly/3uWKcEh
- Dafforn TR, Smith CJ (2004) Natively unfolded domains in endocytosis: hooks, lines and linkers. EMBO Rep 5: 1046-1052. Link: https://bit.ly/3oUid4g
- Dyson HJ, Wright PE (2002b) Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol 12: 54-60. Link: https://bit.ly/3rT3atF
- Dyson HJ, Wright PE (2005a) Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6: 197–208. Link: https://bit.ly/3GXTTEK
- Edwards YJ, Lobley AE, Pentony MM, Jones DT (2009) Insights into the regulation of intrinsically disordered proteins in the human proteome by analyzing sequence and gene expression data. Genome Bioogy 10: 1-8. Link: https://bit.ly/3GVfM7Y
- Favorov MO, Kosoy MY, Tsarev SA, Childs JE, Margolis HS (2000) Prevalence of antibody to hepatitis E virus among rodents in the United States. J Infect Dis 181: 449-455. Link: https://bit.ly/3I0qgUE
- Gadhave K, Gehi BR, Kumar P, Xue B, Uversky VN, et al. (2020) The dark side of Alzheimer’s disease: unstructured biology of proteins from the amyloid cascade signaling pathway. Cell Mol Life Sci 77: 4163-4208. Link: https://bit.ly/3rXzE6a
- Giri R, Kumar D, Sharma N, Uversky VN (2016) Intrinsically disordered side of the Zika virus proteome. Front Cell Infect Microbiol 6: 144. Link: https://bit.ly/3LFT5bu
- Gsponer J, Futschik ME, Teichmann SA, Babu MM (2008) Tight regulation of unstructured proteins: from transcript synthesis to protein degradation. Science 322: 1365–1368. Link: https://bit.ly/3sHBR4G
- Johne R, Heckel G, Plenge-Bönig A, Kindler E, Maresch C, et al. (2010) Novel hepatitis E virus genotype in Norway rats, Germany. Emerg Infect Dis 16:1452-1455. Link: https://bit.ly/3BtD6s9
- Kabrane-Lazizi Y, Fine JB, Elm J, Glass GE, Higa H, et al. (1999) Evidence for widespread infection of wild rats with hepatitis E virus in the United States. Am J Trop Med Hyg 61: 331-335. Link: https://bit.ly/3v0IiT9
- Kenney SP, Meng XJ (2019) Hepatitis E virus genome structure and replication strategy. Cold Spring Harb Perspect Med 9: a031724. Link: https://bit.ly/3rV2cwT
- Khuroo MS, Khuroo MS (2016) Hepatitis E: an emerging global disease–from discovery towards control and cure. J Viral Hepat 23: 68-79. Link: https://bit.ly/3rRxC7s
- Kumar S, Subhadra S, Singh B, Panda BK (2013) Hepatitis E virus: the current scenario. Int J Infect Dis 17: e228-233. Link: https://bit.ly/3JvYTlO
- Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, et al. (2003) Protein disorder prediction: implications for structural proteomics. Structure 11: 1453-1459. Link: https://bit.ly/36kXfFs
- Macdonald A, Harris M (2004) Hepatitis C virus NS5A: tales of a promiscuous protein. J Gen Virol 85: 2485–2502. Link: https://bit.ly/3rUxIuS
- Meng XJ (2010) Hepatitis E virus: animal reservoirs and zoonotic risk. Vet Microbiol 140: 256-265. Link: https://bit.ly/3BpPcTj
- Mishra PM, Verma NC, Rao C, Uversky VN, Nandi CK (2020) Intrinsically disordered proteins of viruses: involvement in the mechanism of cell regulation and pathogenesis. Prog Mol Biol Transl Sci 174: 1-78. Link: https://bit.ly/3gQ80Bp
- Oldfeld CJ, Dunker AK (2014) Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem 83: 553–584. Link: https://bit.ly/3Bq6PlG
- Purdy MA, Lara J, Khudyakov YE (2012) The hepatitis E virus polyproline region is involved in viral adaptation. PLoS One 7: e35974. Link: https://bit.ly/36l6YLX
- Rizzetto M (2009) Hepatitis D: thirty years after. J Hepatol 50: 1043–1050. Link: https://bit.ly/3oUy02W
- Schielke A, Sachs K, Lierz M, Appel B, Jansen A, et al. (2009) Detection of hepatitis E virus in wild boars of rural and urban regions in Germany and whole genome characterization of an endemic strain. Virol J 6: 1-7. Link: https://bit.ly/3oQ6rIj
- Shafat Z, Ahmed A, Parvez MK, Parveen S (2021a) Role of “dual-personality” fragments in HEV adaptation—analysis of Y-domain region. J Genet Eng Biotechnol 19: 1-21. Link: https://bit.ly/3v4j2eC
- Shafat Z, Ahmed A, Parvez MK, Parveen S (2021b) Role of ORF4 in Hepatitis E virus regulation: analysis of intrinsically disordered regions. Journal of Proteins & Proteomics 12. Link: https://bit.ly/3sLIWBo
- Shafat Z, Ahmed A, Parvez MK, Parveen S (2021c) Sequence to structure analysis of the ORF4 protein from Hepatitis E virus. Bioinformation 17: 818-828. Link: https://bit.ly/350zWQw
- Shafat Z, Ahmed A, Parvez MK, Parveen S (2021d) Shedding light on the dark proteome of Hepatitis E Virus. Network Biology 11: 295-314. Link: https://bit.ly/3sJNsjD
- Singh A, Kumar A, Yadav R, Uversky VN, Giri R (2018) Deciphering the dark proteome of Chikungunya virus. Scientific Reports 8: 5822. Link: https://go.nature.com/34OlfjR
- Takahashi M, Tanaka T, Takahashi H, Hoshino Y, Nagashima S, et al. (2010) Hepatitis E Virus (HEV) strains in serum samples can replicate efficiently in cultured cells despite the coexistence of HEV antibodies: characterization of HEV virions in blood circulation. J Clin Microbiol 48: 1112-1125. Link: https://bit.ly/3JQn1jt
- Tam AW, Smith MM, Guerra ME, Huang CC, Bradley DW, et al. (1991) Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology 185: 120-131. Link: https://bit.ly/3uU7UkH
- Uversky VN (2011) Intrinsically disordered proteins from A to Z. Int J Biochem Cell Biol 43: 1090-1103. Link: https://bit.ly/36acvom
- Van Der Lee R, Buljan M, Lang B, Weatheritt RJ, Daughdrill GW, et al. (2014) Classification of intrinsically disordered regions and proteins. Chem Rev 114: 6589–6631. Link: https://bit.ly/3JQnbaz
- Van der Lee R, Lang B, Kruse K, Gsponer J, de Groot NS, et al. (2014) Intrinsically disordered segments affect protein half-life in the cell and during evolution. Cell Reports 8: 1832-1844. Link: https://bit.ly/3v27etH
- Verdegem D, Badillo A, Wieruszeski JM (2011) Domain 3 of NS5A protein from the hepatitis C virus has intrinsic α-helical propensity and is a substrate of cyclophilin A. J Biol Chem 286: 20441–20454. Link: https://bit.ly/3oXspJv
- Wang J, Cao Z, Zhao L, Li S (2011) Novel strategies for drug discovery based on intrinsically disordered proteins (IDPs). Int J Mol Sci 12: 3205–3219. Link: https://bit.ly/3H0sVMS
- Wright PE, Dyson HJ (1999a) Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. Journal of Molecular Biology 293: 321–331. Link: https://bit.ly/3GPwhlS
- Wright PE, Dyson HJ (2015b) Intrinsically disordered proteins in cellular signaling and regulation. Nat Rev Mol Cell Biol 16: 18-29. Link: https://bit.ly/3rVYD9W
- Xue B, Blocquel D, Habchi J, Uversky AV, Kurgan L, et al. (2014) Structural disorder in viral proteins. Chem Rev 114: 6880-91119. Link: https://bit.ly/356kp1L
- Xue B, Williams RW, Oldfield CJ, Goh GK, Dunker AK, et al. (2010) Viral disorder or disordered viruses: do viral proteins possess unique features? Protein Pept Lett 17: 932–951. Link: https://bit.ly/3rRzH3g
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
