Arch Hepat Res
Association of the Treatment Induced Clearance of Hepatitis C Virus Infection with the IL28B Gene Polymorphisms
Pavlina Dzekova Vidimliski1, Igor G Nikolov1, Nadica Matevska Geshkovska2, Yana Boyanova3, Nina Nikolova3, Grigore Romanciuc4, Dan Dumitrascu5, Viktorija Caloska-Ivanova6, Nenad Joksimovic6, Krasimir Antonov3, Lyudmila Mateva3, Lionel Rostaing7, Aleksandar Dimovski2, Aleksandar Sikole1*
2Faculty of Pharmacy, University “Ss Cyril and Methodius”, Skopje, R. Macedonia
3Clinic of Gastroenterology, St Ivan Rilski University Hospital, Sofia, Bulgaria
4Université de Medicine “Nicolae Testemitanu” Chisinau, Moldavie
5University of Medicine and Pharmacy Iuliu Hatieganu, Cluj‑Napoca, Romania
6University Hospital of Gastroenterohepatology, Skopje, R. Macedonia
7Department of Nephrology, Dialysis and Organ Transplantation, CHU Rangueil, Toulouse, France
Cite this as
Vidimliski PD, Nikolov IG, Geshkovska NM, Boyanova Y, Nikolova N, et al. (2015) Association of the Treatment Induced Clearance of Hepatitis C Virus Infection with the IL28B Gene Polymorphisms. Arch Hepat Res 1(1): 005-008. DOI: 10.17352/ahr.000002Background: It has been shown that single nucleotide polymorphisms (SNPs) near the interleukin 28B (IL28B) gene were associated with sustained viral response following standard treatment of hepatitis C virus infection.
Aim: The aim of the study was to evaluate the association between the SNPs near the IL28B gene and the response to the treatment of chronic hepatitis C.
Methods: The genotyping of the three IL28B gene polymorphisms: rs12979860, rs8099917, and rs12980275 was done in 100 Caucasian patients with chronic hepatitis C previously treated with standard antiviral therapy. The study group consisted of 28 hemodialysis patients with end stage renal disease treated with pegylated interferon α and 72 patients without renal disease treated with pegylated interferon α and ribavirin. All patients finished the antiviral treatment at least 6 months before enrollment in the study. Sustained viral response, defined as an absence of detectable HCV RNA in the serum, was tested by an assay with a sensitivity of 20 IU/mL.
Results: Sustained viral response was achieved in 56% (56/100) of the treated patients. The three IL28B gene polymorphisms (CC genotype of rs12979860, TT genotype of rs8099917, and AA genotype of rs12980275) were associated with sustained viral response (p=0.006, p=0.002, p=0.007, respectively) in study patients with chronic hepatitis C treated with standard antiviral therapy.
Conclusion: The IL28B gene polymorphisms: rs12979860, rs8099917, and rs12980275 were significantly associated with the successful treatment of chronic hepatitis C.
Introduction
Hepatitis C virus (HCV) has often been referred to as the “silent virus,” as most HCV infections are clinically silent until the disease reaches a late stage, which often occurs several decades after initial infection. Although a variety of host factors play a role in eradication of HCV, only 15%-25% of adults spontaneously clear the infection [1]. The remaining 75%-85% of patients continue to have persistent viremia, life-long HCV infection, chronic hepatitis C [2].
In the early 2000s, the combination of pegylated interferon alpha (PEGIFNα) and daily doses of ribavirin (RBV) became the standard-of-care (SOC) treatment for chronic hepatitis C [3,4]. The primary goal of the treatment is eradication of HCV, which is synonymous with sustained viral response (SVR), defined as undetectable HCV RNA in blood, 24 weeks after completion of the antiviral treatment [5]. The PEGIFN/RBV treatment is prolonged and expensive, complicated by side effects leading to treatment discontinuation, and only about one-half of the treated patients infected with HCV genotype 1 are achieving sustained viral response [6].
A novel family of antiviral cytokines was described and designated as type III interferons. The interferon type III family consists of three cytokines: Interleukin 29 (interferon lambda 1, IFNλ1), Interleukin 28A (interferon lambda 2, IFNλ2), and Interleukin 28B (interferon lambda 3, IFNλ3). IFNλ is rapidly induced during HCV infection and has antiviral activity against the virus. The type III interferons induce antiviral activity through both innate and adaptive immune pathways [7-9].
The IL28B gene encodes the cytokine interferon lambda 3, IFNλ3. Four genome wide association studies (GWAS) had associated treatment-induced clearance of HCV following PEGIFN/RBV therapy with the single nucleotide polymorphisms (SNPs) near the interleukin 28B (IL28B) gene on chromosome 19 [10-13]. Differences in genetic sequences among individuals are called genetic polymorphisms. The GWAS showed that SNPs near the IL28B gene (CC of rs12979860, TT of rs8099917, and AA of rs12980275) were significantly associated with the successful treatment of chronic hepatitis C. The chance of cure with this standard treatment is twice as high in patients who are homozygous for cytosine in this location (the CC genotype) than in those who are heterozygous (CT) or homozygous for thymine in this location (the TT genotype). The mechanism underlying the association between the IL28B gene polymorphism and response to hepatitis C treatment is not well understood.
The aim of the study was to evaluate the association between single nucleotide polymorphisms near the IL28B gene and the response to the treatment of chronic hepatitis C.
Patients and Methods
Patients
A study group of 100 adult Caucasian patients with chronic hepatitis C routinely treated with antiviral therapy was investigated in the study. The study was approved by the local Ethics Committee and written informed consent was obtained from each study participant. The cohort consisted of 28 patients with end stage renal disease on hemodialysis (HD) treatment and 72 patients without renal disease (non-renal). The HD patients received antiviral therapy only with pegylated interferon α-2a (PEGIFNα-2a). The non-renal patients were treated with pegylated interferon α-2a or α-2b (PEGIFNα-2b) plus ribavirin. The pegylated interferon was administered subcutaneously once a week in a standard dose (PEGIFNα-2a: 135 μg for HD patients and 180 μg for non-renal patients, and PEGIFNα-2b:1.5µg/kg). The ribavirin was administered daily in a dose of 1000 mg for patients with body weight less than 75 kg and 1200 mg for patients with body weight over than 75 kg when was combined with PEG IFNα-2a. The ribavirin dose was 800 mg for patients with body weight less than 65 kg, 1000 mg for patients with body weight 65–85 kg and 1200 mg for patients with body weight over than 85 kg when was combined with PEG IFNα-2b. The treatment duration was 24 weeks for patients infected with HCV genotype 2 and 3, and 48 weeks for the infection caused by HCV genotype 1 and 4. All patients finished the antiviral treatment at least 6 months before enrollment in the study. Sustained viral response (SVR), defined as an absence of detectable HCV RNA in the serum, 6 months after completion of the antiviral treatment, was tested by an assay with a sensitivity of 20 IU/mL.
Methods
Peripheral blood samples in EDTA as an anticoagulant were obtained from each patient enrolled in the study.
HCV quantification
Hepatitis C virus RNA was extracted from plasma using QIAamp Viral RNA kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions. Reverse transcriptase- polymerase chain reaction (RT-PCR) assay for HCV quantification was done with HCV Real-TM Quant (Sacace Biotechnologies, Como, Italy) on Stratagene MX3005P real-time PCR system (Agilent Technologies, Edinburgh, UK) according to manufacturer’s instructions. Detection limit of the assay is 20 IU/ml.
IL28B polymorphisms genotyping
Peripheral blood mononuclear cells (PBMCs) were isolated by density-gradient centrifugation using Histopaque-1077 (Sigma-Aldrich, Munich, Germany) and homogenized in Tri Reagent Solution (Ambion, Life Technologies and Carlsbad, CA, USA). Genomic DNA was extracted from PBMCs homogenized in Tri Reagent Solution (Ambion, Life Technologies, Carlsbad, CA, USA) according to manufacturer’s instructions and genotyped for three IL28B polymorphisms: rs8099917, rs12979860 and rs12980275. The rs8099917 polymorphism was genotyped using TaqMan predesigned SNP genotyping assay (reference C_11710096_10, Applied Biosystems) according manufacturer’s recommended protocol in a total volume of 25µl. The two later SNPs were genotyped using custom designed TaqMan assays with the following primes and probes: TCTACTGAACCAGGGAGCTC, GCGCGGAGTGCAATTCAAC, 6Fam-TGGTTCACGCCTTC, Vic-TGGTTCGCGCCTTC for rs12979860, and GTGCTGAGAGAAGTCAAATTCC, CCGCTACCCGGCAAATATT, 6Fam-ACACGTCCGTTTCTA, Vic-AGACACGTCTGTTTCTA for rs12980275 [14]. For both assays 20ng of DNA was used in a total volume of 25µl including 12,5µl TaqMan Universal PCR master Mix (2x), 1µM of each primer and 200nM of each probe. The PCR reaction conditions were as follows: initial denaturing at 95°C for 10min; 40 cycles of 15 sec at 92°C and 1 min at 64°C to reduce miss priming. Thermal cycling was performed using a Stratagene MX3005P real-time PCR system (Agilent Technologies, Edinburgh, UK). Both positive and negative controls were included in every genotyping assay.
Statistical analysis
Statistical analysis of data was performed using SPSS Statistics 17.0. The parametric variables are presented as the mean and standard deviation. A logistic regression model was used to determine the predictors of the SVR. Odds ratios (ORs) and 95% confidence intervals (95% CIs) were derived from the logistic regression model. The Fisher’s exact test was used to compare proportions. A p value was calculated with two tails. A two-tailed p value of less than or equal to 0.05 was considered statistically significant.
Results
The study group included 100 patients, 71 (71%) men and 29 (29%) women, with a mean age of 43.5 ± 11.4 years. All patients were Caucasians. The HCV genotype 1 caused the chronic hepatitis C infection in most of the patients (93%, 93/100 patients). The HCV genotype 2 and 4 were present in two patients respectively and HCV genotype 3 in one patient. Dual infection with genotype 1 and 3 was also identified in two patients. Sustained viral response was achieved in 56% (56/100) of the treated patients, confirmed by RT-PCR assay with detection limit of 20 IU/ml (Table 1).
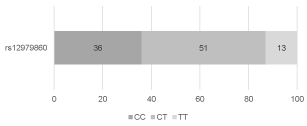
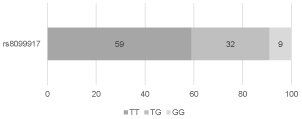
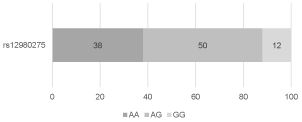
The distribution of the frequencies of rs12979860 genotypes in the study group was as follows: 36 (36%) patients with CC genotype, 51 (51%) patients with CT genotype, and 13 (13%) patients with TT genotype (Figure 1). The distribution of the frequencies of rs8099917 genotypes was: 59 (59%) patients with TT genotype, 32 (32%) patients with TG genotype, and 9 (9%) patients with GG genotype (Figure 2). The distribution of the frequencies of rs12980275 genotypes was: 38 (38%) patients with AA genotype, 50 (50%) patients with AG genotype, and 12 (12%) patients with GG genotype (Figure 3).
The genotype distributions for rs12979860 (CC vs. CT and TT), rs8099917 (TT vs. TG and GG), and rs12980275 (AA vs. AG and GG) polymorphisms were significantly different between SVR group and non SVR group. The achievement of sustained viral response was significantly higher in patients with CC genotype of rs1297860 than in the patients with non CC genotypes (75% vs. 45.3%, p=0.006), Table 2. The SVR was achieved in 75% of patients with the genotype CC of rs12979860, compared with 45.3% of patients with the genotype CT or TT. The occurrence of SVR was significantly higher in patients with TT genotype of rs8099917 than in patients with non TT genotypes (69.5% vs. 36.6%, p=0.002), Table 2. There was also significant association between SVR and rs12980275, SVR was achieved in 73.7% of patients with AA genotype versus 45.2% of patients with non AA genotypes (p=0.007), Table 2.
Gender (OR 1.17, 95% CI, 0.94-1.45, p=0.153), age (OR 0.99, 95% CI, 0.98-1.00, p=0.098), and HCV genotype (OR 1.46, 95% CI, 0.94-2.28, p=0.098) were not significantly associated with achievement of the SVR.
Discussion
The three most widely studied SNPs near the IL28B gene in the genome wide association studies [10-13], were also identified in our study, to evaluate their association with the treatment-induced clearance of HCV in the patients treated with PEGIFN/RBV. The genome wide association studies identified that the homozygosis for the C allele of rs12979860, the homozygosis for the T allele of rs8099917, and the homozygosis for the A allele of rs12980275 were favorable genotypes which predicted treatment response. The same favorable genotypes were associated with the achievement of sustained viral response in treated patients in our study. Most of the studies investigated the effect of the IL28B gene polymorphisms on the SVR in patients with HCV genotype 1. Almost all of treated patients in our study were infected with HCV genotype 1, only 7% of the patients were infected with genotypes 2, 3 and 4. The study of Eslam M et al. [15], confirmed similar effects of the IL28B gene polymorphisms on the SVR in Caucasian patients infected with HCV genotypes 2, 3 and 4 as in the patients with HCV genotype 1.
The distribution of frequencies of rs12979860 genotypes in our study group was: CC in 36%, CT in 51% and TT in 13% of the participants. In Latvia, majority of the people are Caucasians, and they reported similar distribution of frequencies of rs12979860: CC in 33%, CT in 53% and TT in 14% of study participants (Caucasians, n=142) [16]. Similar to our findings, the distribution of frequencies of rs8099917 genotypes among study participants (Caucasians, n= 175) in the study of Sticchi L et al., was as follows: TT in 55%, TG in 40% and GG in 5% [17].
The achieved SVR in the study was 56% and it was associated with the favorable genotypes of the IL28B gene polymorphisms. Consistent to our findings were the results from the study of Domagalski K et al. [18]. The predictability of the three most widely studied SNPs on the treatment response was determined in the cohort of 174 Caucasian (Polish) patients infected with HCV genotype 1 and 4 treated with PEGIFN/RBV. The CC genotype of rs12979860, TT genotype of rs8099917, and AA genotype of rs12980275 were significantly associated with the successful treatment (p=0.001, p=0.016, and p=0.002, respectively) [18]. Similar SVR rates and association with the IL28B gene polymorphisms were evaluated in the studies of Sporea I et al. [19] and Tolmane I et al. [16], although they studied only the rs12979860 as SNP. The study of Sporea I et al. included a cohort of 107 Caucasian (Romanian) patients infected with HCV genotype 1 treated with PEGIFN/RBV [19]. The SVR rate was 50.5% and there was a significant association between the SVR and CC genotype of rs12979860 (73.1% in the CC genotype vs. 43.7% in the non CC genotypes, p=0.012). The study of Tolmane I et al. included 142 Caucasian (Latvian) patients infected with HCV genotype 1 (61%) and HCV genotype 2 or 3 (39%) treated with PEGIFN/RBV [16]. The SVR rate was 59%, and it was significantly associated with the CC genotype of rs12979860 (74% in the CC genotype vs. 52% in the non CC genotypes, p=0.002).
Conclusions
The IL28B gene polymorphisms are strong predictors of the responsiveness to interferon-based regimen, and their determination has demonstrated a great potential to improve patient care. To date, direct antiviral agents including first and second generation protease inhibitors and the polymerase inhibitors represent the available options for highly effective interferon containing and interferon-free regiments for HCV eradication. In near future, even more direct antiviral agents will be approved for various combinations of interferon-free treatment options. However, in many countries approval and reimbursement of direct antiviral agents will be delayed. Also, PEGIFN/RBV therapy may be cost effective option to achieve an SVR in a subgroup of patients with beneficial genetic predictors of treatment response. Additionally, patients who have contraindications to these newer therapies will still need an interferon-based regimen, and thus the IL28B gene polymorphisms will still be important in predicting treatment response and prognosis.
Authors wish to thank Mihaela Codreanu from Agence Universitaire de la Francophonie (AUF), for her administrative support in organization and realization of the regional multicenter collaboration.
- Seeff LB (1999) Natural history of hepatitis C. Am J Med 107: 10S-15S.
- Alter MJ, Kruszon-Moran D, Nainan OV, McQuillan GM, Gao F, et al. (1999) The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N Engl J Med 341: 556-562 .
- Glue P, Rouzier-Panis R, Raffanel C, Sabo R, Gupta SK, et al. (2000) A dose-ranging study of pegylated interferon alfa-2b and ribavirin in chronic hepatitis C. The Hepatitis C Intervention Therapy Group. Hepatology 32: 647–653 .
- Kronenberger B, Zeuzem S (2009) Current and future treatment options for HCV. Ann Hepatol 8: 103–112.
- Pearlman BL, Traub N (2011) Sustained virologic response to antiviral therapy for chronic hepatitis C virus infection: a cure and so much more. Clin Infect Dis 52: 889-900 .
- Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, et al. (2002) Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med 347: 975–982 .
- Rehermann B (2009) Hepatitis C virus versus innate and adaptive immune responses: a tale of coevolution and coexistence. J Clin Invest 119:1745–1754 .
- Marcello T, Grakoui A, Barba-Spaeth G, Machlin ES, Kotenko SV, et al. (2006) Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology 131: 1887–1898 .
- Doyle SE, Schreckhise H, Khuu-Duong K, Henderson K, Rosler R, et al. (2006) Interleukin-29 uses a type 1 interferon-like program to promote antiviral responses in human hepatocytes. Hepatology 44:896–906 .
- Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, et al. (2009) Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461: 399-401.
- Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, et al. (2009) IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet 41: 1100-1104 .
- Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, et al. (2009) Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet 41:1105-1109 .
- Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, et al. (2010) Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology 138: 1338–1345 .
- Lagging M, Askarieh G, Negro F, Bibert S, Söderholm J, et al. (2011) Response prediction in chronic hepatitis C by assessment of IP-10 and IL28B-related single nucleotide polymorphisms. PLoS One 6: e17232.
- Eslam M, Leung R, Romero-Gomez M, Mangia A, Irving WL, et al. (2014) IFNL3 polymorphisms predict response to therapy in chronic hepatitis C genotype 2/3 infection. J Hepatol 61: 235-241 .
- Tolmane I, Rozentale B, Keiss J, Ivancenko L, Subnikova N, et al. (2012) Interleukin 28B Gene Polymorphism and Association with Chronic Hepatitis C Therapy Results in Latvia. Hepat Res Treat ID324090 .
- Sticchi L, Di Biagio A, Rappazzo E, Setti M, De Rosa G, et al. (2013) Rs12979860 and rs8099917 single nucleotide polymorphisms of interleukin-28B gene: simultaneous genotyping in Caucasian patients infected with hepatitis C virus. J Prev Med Hyg 54: 83-86 .
- Domagalski K, Pawlowska M, Tretyn A, Halota W, Tyczyno M, et al. (2013) Association of IL28B Polymorphisms With the Response to Peginterferon Plus Ribavirin Combined Therapy in Polish Patients Infected With HCV Genotype 1 and 4. Hepat Mon 13: e13678.
- Sporea I, Popescu A, Curescu M, Sirli R, Dan I, et al. (2011) The Correlation of Il28B Genotype With Sustained Virologic Response In Romanian patients With Chronic Hepatitis C. Hepat Mon 11: 975-979 .
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
