Annals of Antivirals and Antiretrovirals
Thiolutin Derivatives as inhibitor of RNA-Dependent RNA Polymerase (RdRP) of Hepatitis C Virus: An In-silico Approach
Muhammad Imran Qadir*, Asma Munawar, Syed Bilal Hussain
Cite this as
Qadir MI, Munawar A, Hussain SB (2018) Thiolutin Derivatives as inhibitor of RNA-Dependent RNA Polymerase (RdRP) of Hepatitis C Virus: An In-silico Approach. Ann Antivir Antiretrovir 2(1): 001-003. DOI: 10.17352/aaa.000003Docking study of few Thiolutin derivatives, which originally act as Hepatitis C Virus RNA Dependent RNA Polymerase (RdRP) inhibitor, was performed by using AutoDock Vina. The docking studies reveal that thiolutin interacted with Hepatitis C Virus RNA Dependent RNA Polymerase through hydrogen bonding as well as hydrophobic interactions. Its binding energy after modification (Thiolutin1) was -6.7 kcal/mol, which was greater than the original thiolutin affinity.
Introduction
Major cause of liver failure and liver transplants is Hepatitis C virus (HCV) [1]. According to an estimate 170 million people have HCV which mostly develops chronic infection that takes them to cirrhosis and hepatocellular carcinoma [2-5]. It is treated by pegylated interferon-alpha (IFNα) along with ribavirin. Through this treatment 80% of patients infected by viruses of genotype 2 and 3 are treated. Only 40% of patients infected by genotype 1 HCV are treated. Some patients have to withdraw from the treatment because these drugs cause potentially severe side effect [6].
The hepatitis virus is a positive RNA virus which has four enzymatic activities including NS2 protease, a polymerase concealed by NS5B, protease and helicase concealed by NS3. For genome replication HCV depends upon RNA dependent RNA polymerase which is an important target for inhibitors development. Because of less dependence of RdRp large number of sequence diversity is present in HCV genome across isolates [7]. It provides a challenge to determine the efficacy of anti-HCV drugs, due to both structural variables and rapid growth of resistance in the primary drug target. To get better insight into the Inhibitory mechanism of Hepatitis C Virus RNA-Dependent RNA Polymerase, a series of Azapteridine derivatives were collected from literature and studied against Hepatitis C Virus RNA-Dependent RNA Polymerase enzyme by molecular docking.
Objective of the present study was evaluation of thiolutin derivatives as inhibitor of RNA-dependent RNA polymerase, of hepatitis C virus by using In-silico approach.
Materials and Methods
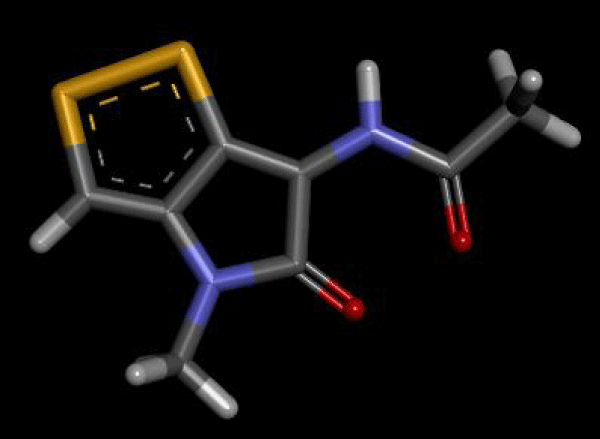
During the molecular docking of the above compound following materials were used in docking: 1) Thiolutin pubchem CID. 6870 2) Hepatitis C Virus RNA Dependent RNA polymerase (RdRP), PDB ID 2gir. 3) m.g.l tools software. 4) Autodock vina. 5) Discovery studio. 6) Pymol. 7) Python. First of all the ligand Thiolutin was downloaded from pubchem.ncbi.org as sdf file (Figure 1).
Then it is opened in discovery studio which was already installed. Hydrogen is added to the ligand to fulfill its valency, then further modification was done to the original ligand molecule and it was saved as PDB file. After that this PDB ligand file is opened in autodock to convert it to PDBQT file to use it in autodock vina. Then target molecule Hepatitis C Virus RNA Dependent RNA Polymerase was downloaded from the RCSB as a text file. This file was opened in discovery studio and ligand which was already present in the protein was selected and deleted to proceed our docking as the already present ligand disturbs the docking with a new ligand. The file was saved as PDB in discovery studio. Then the protein PDB file was opened in autodock and water was deleted and hydrogen was added to the protein molecule. Then the ligand attachment site was selected by making grid box which covered the protein molecule and the values of dimensions and centers were noted. This protein then saved as PDBQT file (Figure 2).
Then ligand and protein files were copied to the vina folder and the values of grid was the conf file was saved and auto docking was started through opening the command prompt. The docking started after command and the program completed docking itself (Figure 3).
Results and Discussion
The dock application is designed to find equipment binding settings between small and medium-sized ligands which are commonly a protein. For each ligand, there are many settings created to try to determine the efforts of binding methods. Docking is beneficial when ligands are flexible molecule. Ligands up to 10 rotatable bands can be properly crafted. The rotating bonds can be used to drive a single-match 3D conformer by using a set of preferred torrent angle. In current docking analysis thiolutin along its derivatives were docked at active site of at Hepatitis C Virus RNA-Dependent RNA Polymerase enzyme. Results of dockings are given as under table 1.
Docking analysis reveal that the most active compound (Thiolutin 1) interacted with receptor active site through polar and hydrophobic interactions, there is hydrogen bond formation reported during docking analysis. The docking results using AutoDock vina is given in the table as binding affinity in kcal/mol. The docking result of Thiolutin showed -6.1 kcal/mol. Then it was modified using Discovery studio changes were made in the original thiolutin and modified it to enhance its binding. Previously, similar compounds like Azapteridine have been proved to inhibitor of RNA-Dependent RNA Polymerase (RdRP) [8].
Binding energy after modification (Thiolutin 1) was -6.7 kcal/mol, which was greater than the original thiolutin affinity.
It is concluded from the present study that Thiolutin after modification (Thiolutin 1), has more binding capacity and may prove useful drug for hepatitis C (Tables 2,3).
- De Francesco R, Tomei L, Altamura S, Summa V, Migliaccio G (2003) Approaching a new era for hepatitis C virus therapy: inhibitors of the NS3-4A serine protease and the NS5B RNA-dependent RNA polymerase. Antiviral Res 58: 1-16. Link: https://goo.gl/u5NLLR
- Bressanelli S, Tomei L, Rey FA, De Francesco R (2002) Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. Virol 76: 3482-3492. Link: https://goo.gl/AUbXhP
- Lesburg CA, Cable MB, Ferrari E, Hong Z, Mannarino AF (1999) Nat Struct Biol 6: 937-943. Manikrao AM, Mahajan NS, Jawarkar RD, Mahajan DT, Masand VH, et al. (2011) J Comput Method Mol Design 1: 35-45. Link: https://goo.gl/g1Nqoq
- Labonte P, Axelrod V, Agarwal A, Aulabaugh A, Amin A, et al. (2002) Modulation of hepatitis C virus RNA-dependent RNA polymerase activity by structure-based site-directed mutagenesis. J Biol Chem 277: 38838-38846. Link: https://goo.gl/FNxinL
- Adachi T, Ago H, Habuka N, Okuda K, Komatsu M, et al. (2002) The essential role of C-terminal residues in regulating the activity of hepatitis C virus RNA-dependent RNA polymerase. Biochim Biophys Acta 1601: 38-48. Link: https://goo.gl/HC1r3M
- Wang M, Ng KK, Cherney MM, Chan L, Yannopoulos CG, et al. (2003) Non-nucleoside analogue inhibitors bind to an allosteric site on HCV NS5B polymerase. Crystal structures and mechanism of inhibition. J Biol Chem 278: 9489-9495. Link: https://goo.gl/uA6uFQ
- Sloley BD, Urichuk LJ, Morley P, Durkin J, Shan JJ, et al. (2000) Identification of kaempferol as a monoamine oxidase inhibitor and potential Neuroprotectant in extracts of Ginkgo biloba leaves. J Pharm Pharmacol 52: 451-459. Link: https://goo.gl/4EB88Q
- T Middleton, HB Lim, D Montgomery, T Rockway, D Liu, et al. (2007) Azapteridine inhibitors of hepatitis C virus RNA-dependent RNA polymerase. Letters in Drug Design & Discovery 4: 1-8. Link: https://goo.gl/gsuYbk
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
